| Full name: limb and CNS expressed 1 like | Alias Symbol: MGC46719 | ||
| Type: protein-coding gene | Cytoband: 1q21.1 | ||
| Entrez ID: 128077 | HGNC ID: HGNC:28715 | Ensembl Gene: ENSG00000271601 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of LIX1L:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LIX1L | 128077 | 225793_at | -0.2213 | 0.7875 | |
| GSE26886 | LIX1L | 128077 | 225793_at | 0.7858 | 0.0075 | |
| GSE45670 | LIX1L | 128077 | 225793_at | -1.0324 | 0.0007 | |
| GSE53622 | LIX1L | 128077 | 13009 | -0.2969 | 0.0243 | |
| GSE53624 | LIX1L | 128077 | 131871 | -0.1763 | 0.0619 | |
| GSE63941 | LIX1L | 128077 | 225793_at | -3.3267 | 0.0073 | |
| GSE77861 | LIX1L | 128077 | 225793_at | 0.2736 | 0.4877 | |
| GSE97050 | LIX1L | 128077 | A_23_P342744 | -0.3840 | 0.2317 | |
| SRP007169 | LIX1L | 128077 | RNAseq | 0.6898 | 0.1459 | |
| SRP008496 | LIX1L | 128077 | RNAseq | 1.3060 | 0.0000 | |
| SRP064894 | LIX1L | 128077 | RNAseq | 0.3873 | 0.0369 | |
| SRP133303 | LIX1L | 128077 | RNAseq | -0.1907 | 0.3489 | |
| SRP159526 | LIX1L | 128077 | RNAseq | -0.1157 | 0.5947 | |
| SRP193095 | LIX1L | 128077 | RNAseq | 0.4094 | 0.0016 | |
| SRP219564 | LIX1L | 128077 | RNAseq | 0.3381 | 0.5449 | |
| TCGA | LIX1L | 128077 | RNAseq | -0.1384 | 0.1137 |
Upregulated datasets: 1; Downregulated datasets: 2.
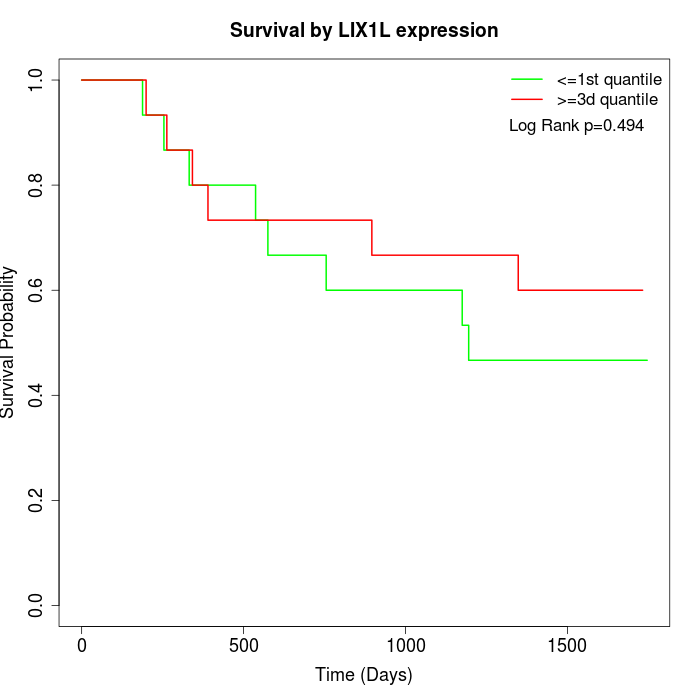
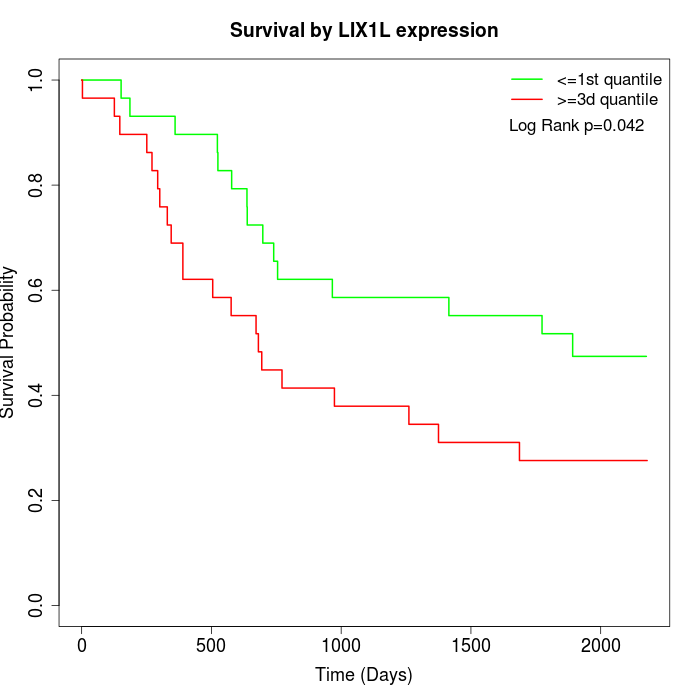
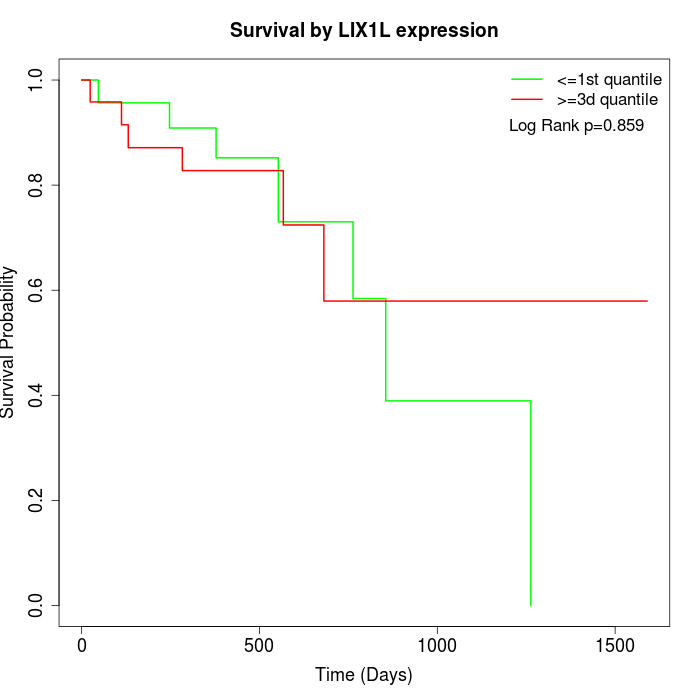
Survival by LIX1L expression:
Note: Click image to view full size file.
Copy number change of LIX1L:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LIX1L | 128077 | 13 | 0 | 17 | |
| GSE20123 | LIX1L | 128077 | 13 | 0 | 17 | |
| GSE43470 | LIX1L | 128077 | 5 | 2 | 36 | |
| GSE46452 | LIX1L | 128077 | 2 | 1 | 56 | |
| GSE47630 | LIX1L | 128077 | 12 | 2 | 26 | |
| GSE54993 | LIX1L | 128077 | 0 | 2 | 68 | |
| GSE54994 | LIX1L | 128077 | 15 | 0 | 38 | |
| GSE60625 | LIX1L | 128077 | 0 | 0 | 11 | |
| GSE74703 | LIX1L | 128077 | 5 | 2 | 29 | |
| GSE74704 | LIX1L | 128077 | 6 | 0 | 14 | |
| TCGA | LIX1L | 128077 | 33 | 15 | 48 |
Total number of gains: 104; Total number of losses: 24; Total Number of normals: 360.
Somatic mutations of LIX1L:
Generating mutation plots.
Highly correlated genes for LIX1L:
Showing top 20/1296 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LIX1L | MCAM | 0.815242 | 6 | 0 | 6 |
| LIX1L | KLF11 | 0.785693 | 4 | 0 | 4 |
| LIX1L | PACS2 | 0.779795 | 3 | 0 | 3 |
| LIX1L | CLMP | 0.778812 | 6 | 0 | 6 |
| LIX1L | DEGS1 | 0.770932 | 3 | 0 | 3 |
| LIX1L | PLOD1 | 0.765559 | 4 | 0 | 4 |
| LIX1L | ANXA6 | 0.765317 | 8 | 0 | 7 |
| LIX1L | MYO5A | 0.765184 | 3 | 0 | 3 |
| LIX1L | DCHS1 | 0.757362 | 6 | 0 | 6 |
| LIX1L | FIBIN | 0.754508 | 5 | 0 | 4 |
| LIX1L | EPHA3 | 0.752435 | 5 | 0 | 5 |
| LIX1L | MAP1A | 0.750846 | 6 | 0 | 6 |
| LIX1L | NENF | 0.747153 | 3 | 0 | 3 |
| LIX1L | GPR3 | 0.747026 | 3 | 0 | 3 |
| LIX1L | STOML1 | 0.746911 | 3 | 0 | 3 |
| LIX1L | ETS2 | 0.745182 | 3 | 0 | 3 |
| LIX1L | ICA1L | 0.743757 | 3 | 0 | 3 |
| LIX1L | ATP8B2 | 0.743371 | 7 | 0 | 6 |
| LIX1L | TCEAL5 | 0.743086 | 3 | 0 | 3 |
| LIX1L | ZCCHC24 | 0.742822 | 8 | 0 | 7 |
For details and further investigation, click here