| Full name: ETS proto-oncogene 2, transcription factor | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 21q22.2 | ||
| Entrez ID: 2114 | HGNC ID: HGNC:3489 | Ensembl Gene: ENSG00000157557 | OMIM ID: 164740 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ETS2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04014 | Ras signaling pathway |
Expression of ETS2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ETS2 | 2114 | 201328_at | -0.2067 | 0.7532 | |
| GSE20347 | ETS2 | 2114 | 201328_at | -0.3314 | 0.2963 | |
| GSE23400 | ETS2 | 2114 | 201328_at | -0.1241 | 0.3337 | |
| GSE26886 | ETS2 | 2114 | 201328_at | 0.3105 | 0.3690 | |
| GSE29001 | ETS2 | 2114 | 201328_at | -0.0537 | 0.8695 | |
| GSE38129 | ETS2 | 2114 | 201328_at | -0.1326 | 0.5913 | |
| GSE45670 | ETS2 | 2114 | 201328_at | 0.1340 | 0.6720 | |
| GSE53622 | ETS2 | 2114 | 120437 | -0.4352 | 0.0003 | |
| GSE53624 | ETS2 | 2114 | 120437 | -0.4406 | 0.0000 | |
| GSE63941 | ETS2 | 2114 | 201329_s_at | 0.0553 | 0.9353 | |
| GSE77861 | ETS2 | 2114 | 201328_at | 0.1116 | 0.7658 | |
| GSE97050 | ETS2 | 2114 | A_24_P314179 | -0.4592 | 0.1324 | |
| SRP007169 | ETS2 | 2114 | RNAseq | -1.6111 | 0.0002 | |
| SRP008496 | ETS2 | 2114 | RNAseq | -0.7622 | 0.0393 | |
| SRP064894 | ETS2 | 2114 | RNAseq | 0.0665 | 0.8194 | |
| SRP133303 | ETS2 | 2114 | RNAseq | 0.2590 | 0.2687 | |
| SRP159526 | ETS2 | 2114 | RNAseq | -0.1232 | 0.7893 | |
| SRP193095 | ETS2 | 2114 | RNAseq | 0.2309 | 0.2428 | |
| SRP219564 | ETS2 | 2114 | RNAseq | 0.8924 | 0.1642 | |
| TCGA | ETS2 | 2114 | RNAseq | 0.0232 | 0.7156 |
Upregulated datasets: 0; Downregulated datasets: 1.
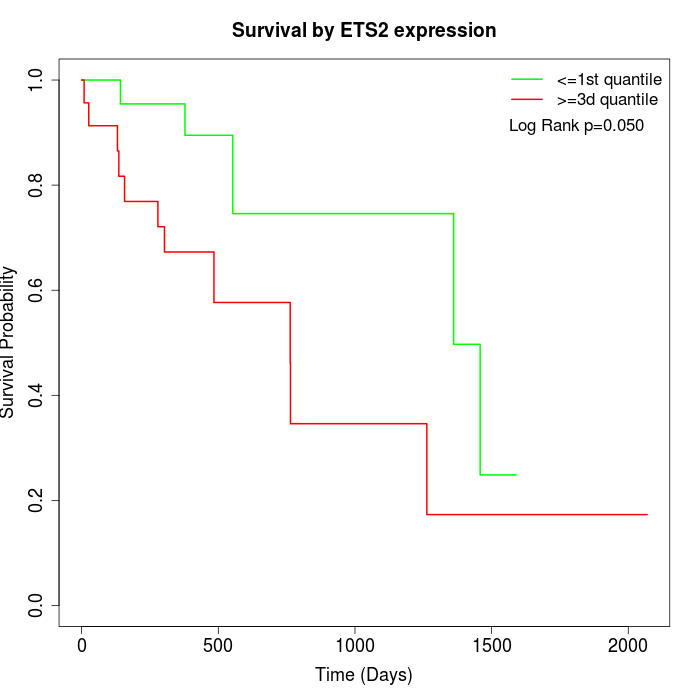
Survival by ETS2 expression:
Note: Click image to view full size file.
Copy number change of ETS2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ETS2 | 2114 | 2 | 8 | 20 | |
| GSE20123 | ETS2 | 2114 | 2 | 8 | 20 | |
| GSE43470 | ETS2 | 2114 | 0 | 10 | 33 | |
| GSE46452 | ETS2 | 2114 | 1 | 21 | 37 | |
| GSE47630 | ETS2 | 2114 | 6 | 17 | 17 | |
| GSE54993 | ETS2 | 2114 | 8 | 1 | 61 | |
| GSE54994 | ETS2 | 2114 | 3 | 9 | 41 | |
| GSE60625 | ETS2 | 2114 | 0 | 0 | 11 | |
| GSE74703 | ETS2 | 2114 | 0 | 8 | 28 | |
| GSE74704 | ETS2 | 2114 | 1 | 5 | 14 | |
| TCGA | ETS2 | 2114 | 9 | 41 | 46 |
Total number of gains: 32; Total number of losses: 128; Total Number of normals: 328.
Somatic mutations of ETS2:
Generating mutation plots.
Highly correlated genes for ETS2:
Showing top 20/127 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ETS2 | NEK9 | 0.796683 | 3 | 0 | 3 |
| ETS2 | UBR1 | 0.780961 | 3 | 0 | 3 |
| ETS2 | TSPAN2 | 0.769833 | 3 | 0 | 3 |
| ETS2 | PLEKHA8 | 0.747763 | 3 | 0 | 3 |
| ETS2 | LIX1L | 0.745182 | 3 | 0 | 3 |
| ETS2 | CYTH3 | 0.740956 | 3 | 0 | 3 |
| ETS2 | ANKMY2 | 0.738477 | 3 | 0 | 3 |
| ETS2 | PAK2 | 0.734192 | 3 | 0 | 3 |
| ETS2 | MRPS27 | 0.728547 | 3 | 0 | 3 |
| ETS2 | PPP3CC | 0.70657 | 3 | 0 | 3 |
| ETS2 | KCTD12 | 0.702119 | 3 | 0 | 3 |
| ETS2 | RPP14 | 0.695478 | 3 | 0 | 3 |
| ETS2 | LYSMD2 | 0.692146 | 3 | 0 | 3 |
| ETS2 | DENND5A | 0.691402 | 3 | 0 | 3 |
| ETS2 | SAMD8 | 0.690406 | 3 | 0 | 3 |
| ETS2 | ASCC3 | 0.689185 | 3 | 0 | 3 |
| ETS2 | FKBP9 | 0.688688 | 4 | 0 | 3 |
| ETS2 | DLL1 | 0.687016 | 3 | 0 | 3 |
| ETS2 | HDDC3 | 0.68585 | 3 | 0 | 3 |
| ETS2 | SIN3A | 0.682083 | 3 | 0 | 3 |
For details and further investigation, click here