| Full name: EPH receptor A3 | Alias Symbol: HEK|HEK4 | ||
| Type: protein-coding gene | Cytoband: 3p11.1 | ||
| Entrez ID: 2042 | HGNC ID: HGNC:3387 | Ensembl Gene: ENSG00000044524 | OMIM ID: 179611 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
EPHA3 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04360 | Axon guidance |
Expression of EPHA3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EPHA3 | 2042 | 206070_s_at | -1.0041 | 0.4215 | |
| GSE20347 | EPHA3 | 2042 | 206070_s_at | -0.0277 | 0.8821 | |
| GSE23400 | EPHA3 | 2042 | 206071_s_at | -0.0464 | 0.1094 | |
| GSE26886 | EPHA3 | 2042 | 206071_s_at | 0.0548 | 0.6626 | |
| GSE29001 | EPHA3 | 2042 | 206071_s_at | -0.1305 | 0.3336 | |
| GSE38129 | EPHA3 | 2042 | 206070_s_at | -0.6684 | 0.0673 | |
| GSE45670 | EPHA3 | 2042 | 206070_s_at | -2.4341 | 0.0000 | |
| GSE53622 | EPHA3 | 2042 | 8582 | -0.7556 | 0.0006 | |
| GSE53624 | EPHA3 | 2042 | 8582 | -0.4787 | 0.0345 | |
| GSE63941 | EPHA3 | 2042 | 206071_s_at | 0.1315 | 0.2757 | |
| GSE77861 | EPHA3 | 2042 | 206071_s_at | -0.0535 | 0.7103 | |
| GSE97050 | EPHA3 | 2042 | A_23_P169819 | -0.5720 | 0.1123 | |
| SRP007169 | EPHA3 | 2042 | RNAseq | -0.3525 | 0.5550 | |
| SRP008496 | EPHA3 | 2042 | RNAseq | 0.7923 | 0.0548 | |
| SRP064894 | EPHA3 | 2042 | RNAseq | 0.2435 | 0.5814 | |
| SRP133303 | EPHA3 | 2042 | RNAseq | -0.1322 | 0.7325 | |
| SRP159526 | EPHA3 | 2042 | RNAseq | -0.0214 | 0.9658 | |
| SRP193095 | EPHA3 | 2042 | RNAseq | 0.2664 | 0.4033 | |
| SRP219564 | EPHA3 | 2042 | RNAseq | 0.7425 | 0.3958 | |
| TCGA | EPHA3 | 2042 | RNAseq | -0.5570 | 0.0103 |
Upregulated datasets: 0; Downregulated datasets: 1.
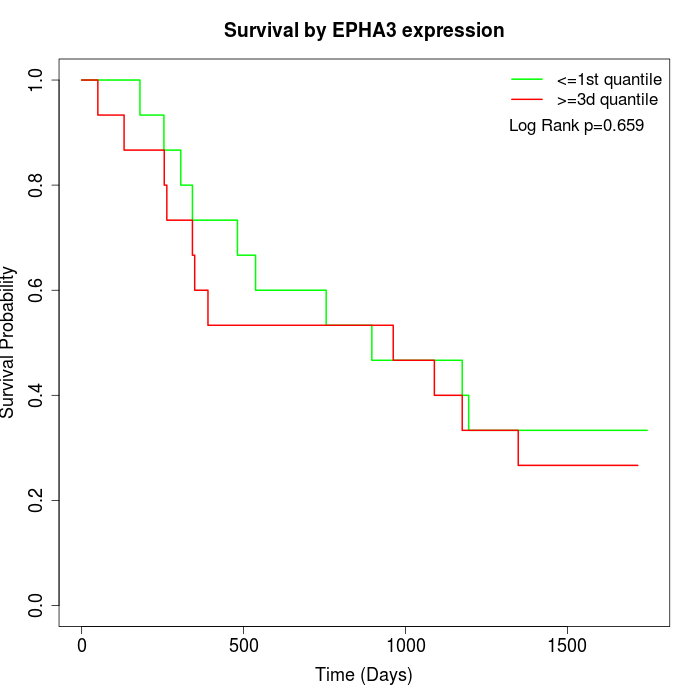
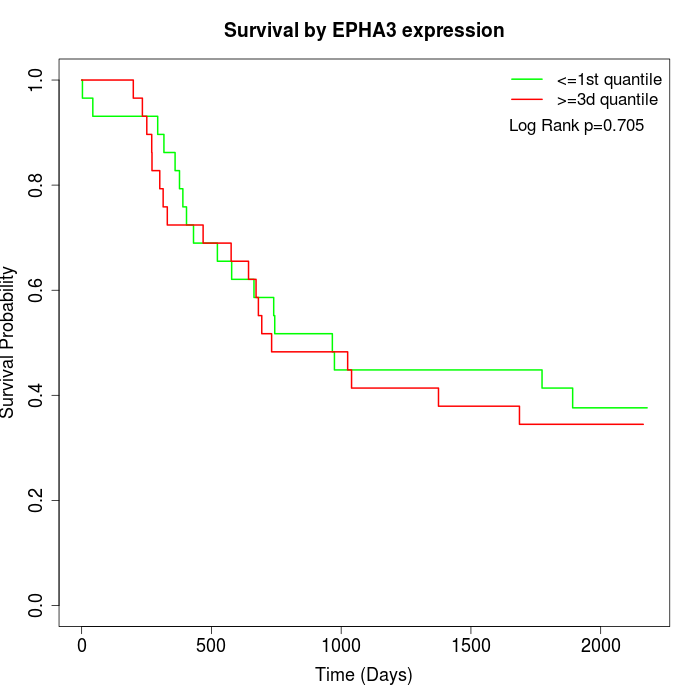
Survival by EPHA3 expression:
Note: Click image to view full size file.
Copy number change of EPHA3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EPHA3 | 2042 | 3 | 15 | 12 | |
| GSE20123 | EPHA3 | 2042 | 3 | 15 | 12 | |
| GSE43470 | EPHA3 | 2042 | 4 | 19 | 20 | |
| GSE46452 | EPHA3 | 2042 | 4 | 16 | 39 | |
| GSE47630 | EPHA3 | 2042 | 6 | 16 | 18 | |
| GSE54993 | EPHA3 | 2042 | 4 | 3 | 63 | |
| GSE54994 | EPHA3 | 2042 | 4 | 26 | 23 | |
| GSE60625 | EPHA3 | 2042 | 0 | 6 | 5 | |
| GSE74703 | EPHA3 | 2042 | 4 | 17 | 15 | |
| GSE74704 | EPHA3 | 2042 | 2 | 10 | 8 | |
| TCGA | EPHA3 | 2042 | 6 | 65 | 25 |
Total number of gains: 40; Total number of losses: 208; Total Number of normals: 240.
Somatic mutations of EPHA3:
Generating mutation plots.
Highly correlated genes for EPHA3:
Showing top 20/800 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EPHA3 | RERE | 0.793336 | 4 | 0 | 4 |
| EPHA3 | CYP2U1 | 0.79328 | 5 | 0 | 5 |
| EPHA3 | SOBP | 0.783255 | 6 | 0 | 6 |
| EPHA3 | NAP1L3 | 0.781542 | 6 | 0 | 6 |
| EPHA3 | GNG2 | 0.775736 | 4 | 0 | 4 |
| EPHA3 | TNFSF12 | 0.771921 | 3 | 0 | 3 |
| EPHA3 | PLCL1 | 0.770601 | 7 | 0 | 7 |
| EPHA3 | CD99L2 | 0.769904 | 3 | 0 | 3 |
| EPHA3 | PLN | 0.764993 | 7 | 0 | 7 |
| EPHA3 | AFF1 | 0.764891 | 3 | 0 | 3 |
| EPHA3 | CNN1 | 0.763015 | 5 | 0 | 4 |
| EPHA3 | NR2F1 | 0.761128 | 3 | 0 | 3 |
| EPHA3 | SLIT2 | 0.76062 | 7 | 0 | 7 |
| EPHA3 | LRCH2 | 0.759578 | 5 | 0 | 5 |
| EPHA3 | C3orf70 | 0.759533 | 5 | 0 | 5 |
| EPHA3 | PCDH9 | 0.755062 | 6 | 0 | 5 |
| EPHA3 | SSPN | 0.754503 | 6 | 0 | 6 |
| EPHA3 | FRZB | 0.753875 | 8 | 0 | 8 |
| EPHA3 | LIX1L | 0.752435 | 5 | 0 | 5 |
| EPHA3 | CCDC30 | 0.750252 | 3 | 0 | 3 |
For details and further investigation, click here