| Full name: LIM domain only 1 | Alias Symbol: TTG1|RHOM1 | ||
| Type: protein-coding gene | Cytoband: 11p15.4 | ||
| Entrez ID: 4004 | HGNC ID: HGNC:6641 | Ensembl Gene: ENSG00000166407 | OMIM ID: 186921 |
| Drug and gene relationship at DGIdb | |||
Expression of LMO1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LMO1 | 4004 | 206718_at | 0.1652 | 0.5878 | |
| GSE20347 | LMO1 | 4004 | 206718_at | 0.1505 | 0.1237 | |
| GSE23400 | LMO1 | 4004 | 206718_at | 0.0522 | 0.1376 | |
| GSE26886 | LMO1 | 4004 | 206718_at | 0.3832 | 0.0009 | |
| GSE29001 | LMO1 | 4004 | 206718_at | 0.0032 | 0.9874 | |
| GSE38129 | LMO1 | 4004 | 206718_at | 0.1068 | 0.2250 | |
| GSE45670 | LMO1 | 4004 | 206718_at | 0.3995 | 0.0019 | |
| GSE63941 | LMO1 | 4004 | 206718_at | 0.3241 | 0.3039 | |
| GSE77861 | LMO1 | 4004 | 206718_at | 0.0033 | 0.9880 | |
| GSE97050 | LMO1 | 4004 | A_23_P87310 | -0.0802 | 0.7207 | |
| TCGA | LMO1 | 4004 | RNAseq | 2.8361 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 0.
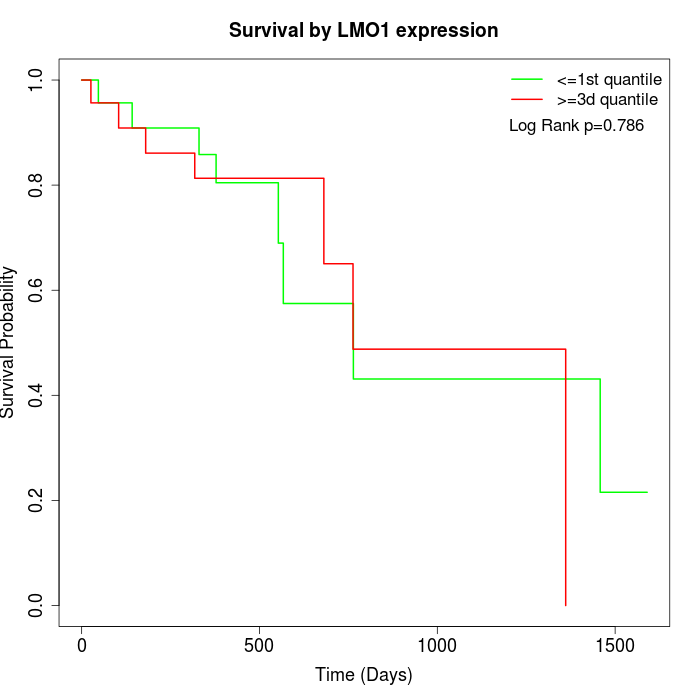
Survival by LMO1 expression:
Note: Click image to view full size file.
Copy number change of LMO1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LMO1 | 4004 | 0 | 11 | 19 | |
| GSE20123 | LMO1 | 4004 | 0 | 11 | 19 | |
| GSE43470 | LMO1 | 4004 | 2 | 5 | 36 | |
| GSE46452 | LMO1 | 4004 | 7 | 5 | 47 | |
| GSE47630 | LMO1 | 4004 | 2 | 12 | 26 | |
| GSE54993 | LMO1 | 4004 | 3 | 1 | 66 | |
| GSE54994 | LMO1 | 4004 | 2 | 12 | 39 | |
| GSE60625 | LMO1 | 4004 | 0 | 0 | 11 | |
| GSE74703 | LMO1 | 4004 | 1 | 4 | 31 | |
| GSE74704 | LMO1 | 4004 | 0 | 9 | 11 | |
| TCGA | LMO1 | 4004 | 9 | 36 | 51 |
Total number of gains: 26; Total number of losses: 106; Total Number of normals: 356.
Somatic mutations of LMO1:
Generating mutation plots.
Highly correlated genes for LMO1:
Showing top 20/445 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LMO1 | PHC2 | 0.832567 | 3 | 0 | 3 |
| LMO1 | NFATC1 | 0.819813 | 3 | 0 | 3 |
| LMO1 | PITPNC1 | 0.805486 | 3 | 0 | 3 |
| LMO1 | XAB2 | 0.783254 | 3 | 0 | 3 |
| LMO1 | SIDT2 | 0.77366 | 3 | 0 | 3 |
| LMO1 | ADAM19 | 0.770502 | 3 | 0 | 3 |
| LMO1 | RILPL2 | 0.768485 | 3 | 0 | 3 |
| LMO1 | PLOD3 | 0.763089 | 3 | 0 | 3 |
| LMO1 | TNS3 | 0.760236 | 3 | 0 | 3 |
| LMO1 | CATSPER1 | 0.755822 | 4 | 0 | 4 |
| LMO1 | HCFC1R1 | 0.754272 | 3 | 0 | 3 |
| LMO1 | ATP9A | 0.745079 | 3 | 0 | 3 |
| LMO1 | TRIM47 | 0.744022 | 3 | 0 | 3 |
| LMO1 | HNRNPUL1 | 0.741137 | 3 | 0 | 3 |
| LMO1 | CERK | 0.739777 | 3 | 0 | 3 |
| LMO1 | PSKH1 | 0.73817 | 3 | 0 | 3 |
| LMO1 | TIMP1 | 0.736549 | 3 | 0 | 3 |
| LMO1 | PIN4 | 0.735196 | 3 | 0 | 3 |
| LMO1 | ASB2 | 0.735066 | 3 | 0 | 3 |
| LMO1 | CACNA1B | 0.730496 | 4 | 0 | 3 |
For details and further investigation, click here