| Full name: ceramide kinase | Alias Symbol: hCERK|FLJ23239|dA59H18.3|DKFZp434E0211|FLJ21430|KIAA1646|LK4|dA59H18.2 | ||
| Type: protein-coding gene | Cytoband: 22q13.31 | ||
| Entrez ID: 64781 | HGNC ID: HGNC:19256 | Ensembl Gene: ENSG00000100422 | OMIM ID: 610307 |
| Drug and gene relationship at DGIdb | |||
Expression of CERK:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CERK | 64781 | 218421_at | 0.2115 | 0.7711 | |
| GSE20347 | CERK | 64781 | 218421_at | 0.2356 | 0.1071 | |
| GSE23400 | CERK | 64781 | 218421_at | 0.1493 | 0.0293 | |
| GSE26886 | CERK | 64781 | 218421_at | 0.4554 | 0.0841 | |
| GSE29001 | CERK | 64781 | 218421_at | -0.0307 | 0.9088 | |
| GSE38129 | CERK | 64781 | 218421_at | 0.1386 | 0.3949 | |
| GSE45670 | CERK | 64781 | 218421_at | 0.1493 | 0.4407 | |
| GSE53622 | CERK | 64781 | 96047 | -0.1428 | 0.1245 | |
| GSE53624 | CERK | 64781 | 96047 | 0.2240 | 0.0182 | |
| GSE63941 | CERK | 64781 | 218421_at | -1.3386 | 0.0338 | |
| GSE77861 | CERK | 64781 | 218421_at | 0.6136 | 0.0706 | |
| GSE97050 | CERK | 64781 | A_24_P62237 | 0.2155 | 0.4357 | |
| SRP007169 | CERK | 64781 | RNAseq | 0.2383 | 0.5262 | |
| SRP008496 | CERK | 64781 | RNAseq | 0.2283 | 0.3753 | |
| SRP064894 | CERK | 64781 | RNAseq | 0.2020 | 0.1534 | |
| SRP133303 | CERK | 64781 | RNAseq | 0.2050 | 0.3453 | |
| SRP159526 | CERK | 64781 | RNAseq | -0.0677 | 0.7602 | |
| SRP193095 | CERK | 64781 | RNAseq | 0.0901 | 0.4310 | |
| SRP219564 | CERK | 64781 | RNAseq | -0.2667 | 0.3585 | |
| TCGA | CERK | 64781 | RNAseq | -0.2192 | 0.0003 |
Upregulated datasets: 0; Downregulated datasets: 1.
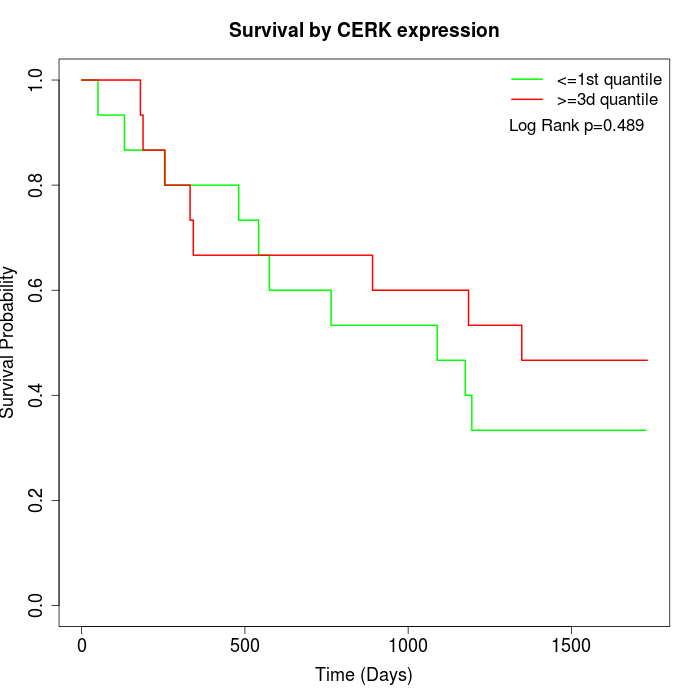
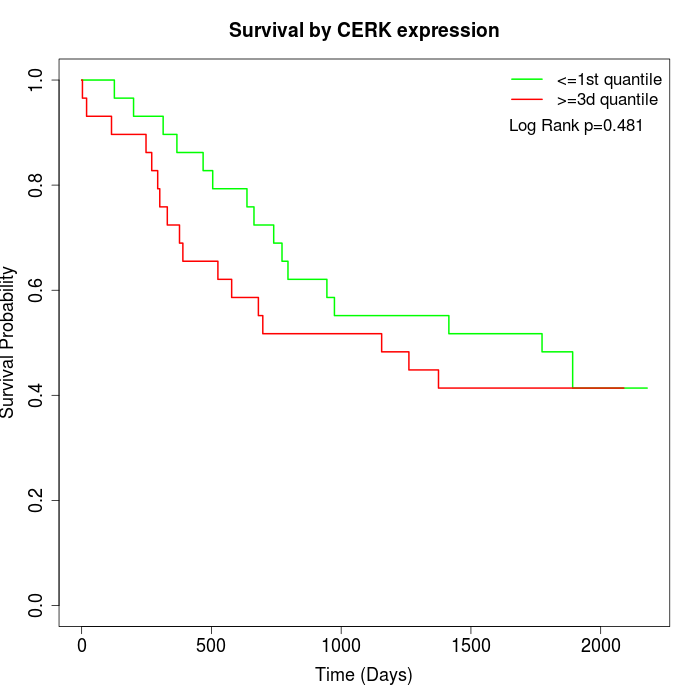
Survival by CERK expression:
Note: Click image to view full size file.
Copy number change of CERK:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CERK | 64781 | 5 | 6 | 19 | |
| GSE20123 | CERK | 64781 | 4 | 5 | 21 | |
| GSE43470 | CERK | 64781 | 2 | 6 | 35 | |
| GSE46452 | CERK | 64781 | 31 | 2 | 26 | |
| GSE47630 | CERK | 64781 | 9 | 4 | 27 | |
| GSE54993 | CERK | 64781 | 3 | 6 | 61 | |
| GSE54994 | CERK | 64781 | 11 | 7 | 35 | |
| GSE60625 | CERK | 64781 | 5 | 0 | 6 | |
| GSE74703 | CERK | 64781 | 2 | 4 | 30 | |
| GSE74704 | CERK | 64781 | 2 | 3 | 15 | |
| TCGA | CERK | 64781 | 27 | 17 | 52 |
Total number of gains: 101; Total number of losses: 60; Total Number of normals: 327.
Somatic mutations of CERK:
Generating mutation plots.
Highly correlated genes for CERK:
Showing top 20/497 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CERK | NUDC | 0.761895 | 3 | 0 | 3 |
| CERK | NAE1 | 0.761698 | 3 | 0 | 3 |
| CERK | PROK2 | 0.753452 | 3 | 0 | 3 |
| CERK | LMO1 | 0.739777 | 3 | 0 | 3 |
| CERK | G0S2 | 0.725583 | 3 | 0 | 3 |
| CERK | TLE1 | 0.723269 | 3 | 0 | 3 |
| CERK | BCL2A1 | 0.719416 | 4 | 0 | 3 |
| CERK | USP28 | 0.712433 | 3 | 0 | 3 |
| CERK | NOA1 | 0.71081 | 3 | 0 | 3 |
| CERK | NAA40 | 0.702433 | 3 | 0 | 3 |
| CERK | SEC24D | 0.701305 | 4 | 0 | 4 |
| CERK | CSNK2A2 | 0.699441 | 3 | 0 | 3 |
| CERK | ABHD2 | 0.699277 | 4 | 0 | 4 |
| CERK | ABCB10 | 0.69895 | 3 | 0 | 3 |
| CERK | STRAP | 0.693789 | 4 | 0 | 3 |
| CERK | ZNF451 | 0.693283 | 3 | 0 | 3 |
| CERK | CBFB | 0.683508 | 4 | 0 | 4 |
| CERK | TTC30B | 0.682399 | 3 | 0 | 3 |
| CERK | KCNE3 | 0.679419 | 3 | 0 | 3 |
| CERK | TUBG2 | 0.676772 | 3 | 0 | 3 |
For details and further investigation, click here