| Full name: MCL1 apoptosis regulator, BCL2 family member | Alias Symbol: BCL2L3|Mcl-1 | ||
| Type: protein-coding gene | Cytoband: 1q21.2 | ||
| Entrez ID: 4170 | HGNC ID: HGNC:6943 | Ensembl Gene: ENSG00000143384 | OMIM ID: 159552 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
MCL1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa04210 | Apoptosis | |
| hsa04630 | Jak-STAT signaling pathway |
Expression of MCL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MCL1 | 4170 | 200797_s_at | -0.1242 | 0.8094 | |
| GSE20347 | MCL1 | 4170 | 200797_s_at | -0.1643 | 0.2593 | |
| GSE23400 | MCL1 | 4170 | 200797_s_at | -0.1574 | 0.0137 | |
| GSE26886 | MCL1 | 4170 | 200797_s_at | -0.0572 | 0.7710 | |
| GSE29001 | MCL1 | 4170 | 200797_s_at | -0.1173 | 0.6112 | |
| GSE38129 | MCL1 | 4170 | 200797_s_at | -0.1328 | 0.2825 | |
| GSE45670 | MCL1 | 4170 | 200797_s_at | -0.1460 | 0.2863 | |
| GSE53622 | MCL1 | 4170 | 54882 | -0.3199 | 0.0000 | |
| GSE53624 | MCL1 | 4170 | 54882 | -0.3934 | 0.0000 | |
| GSE63941 | MCL1 | 4170 | 200797_s_at | -0.4263 | 0.1743 | |
| GSE77861 | MCL1 | 4170 | 200797_s_at | 0.0864 | 0.6510 | |
| GSE97050 | MCL1 | 4170 | A_24_P336759 | -0.0589 | 0.8194 | |
| SRP007169 | MCL1 | 4170 | RNAseq | -0.7191 | 0.0266 | |
| SRP008496 | MCL1 | 4170 | RNAseq | -0.4222 | 0.0583 | |
| SRP064894 | MCL1 | 4170 | RNAseq | -0.5081 | 0.0161 | |
| SRP133303 | MCL1 | 4170 | RNAseq | -0.1055 | 0.5140 | |
| SRP159526 | MCL1 | 4170 | RNAseq | -0.3360 | 0.1728 | |
| SRP193095 | MCL1 | 4170 | RNAseq | -0.0964 | 0.4993 | |
| SRP219564 | MCL1 | 4170 | RNAseq | -0.1392 | 0.6396 | |
| TCGA | MCL1 | 4170 | RNAseq | -0.0464 | 0.2915 |
Upregulated datasets: 0; Downregulated datasets: 0.
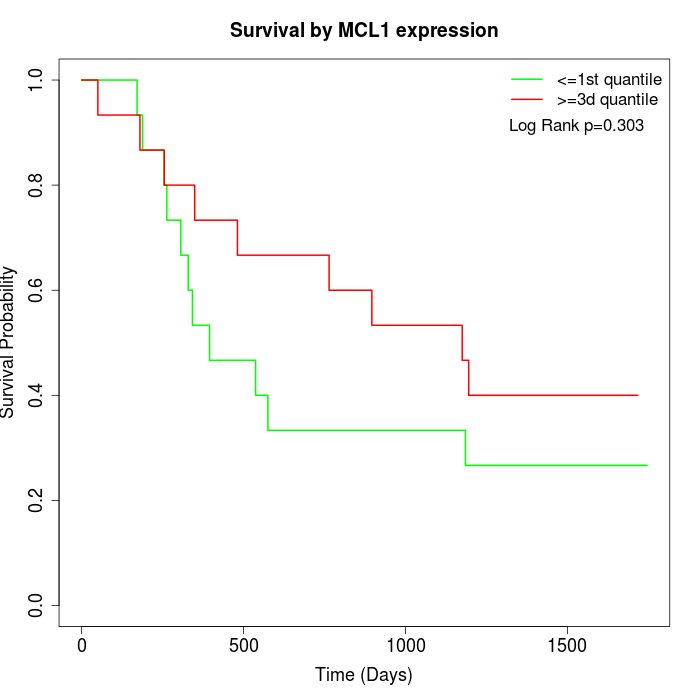
Survival by MCL1 expression:
Note: Click image to view full size file.
Copy number change of MCL1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MCL1 | 4170 | 15 | 0 | 15 | |
| GSE20123 | MCL1 | 4170 | 15 | 0 | 15 | |
| GSE43470 | MCL1 | 4170 | 6 | 1 | 36 | |
| GSE46452 | MCL1 | 4170 | 2 | 1 | 56 | |
| GSE47630 | MCL1 | 4170 | 14 | 0 | 26 | |
| GSE54993 | MCL1 | 4170 | 0 | 3 | 67 | |
| GSE54994 | MCL1 | 4170 | 15 | 0 | 38 | |
| GSE60625 | MCL1 | 4170 | 0 | 0 | 11 | |
| GSE74703 | MCL1 | 4170 | 6 | 1 | 29 | |
| GSE74704 | MCL1 | 4170 | 7 | 0 | 13 | |
| TCGA | MCL1 | 4170 | 38 | 2 | 56 |
Total number of gains: 118; Total number of losses: 8; Total Number of normals: 362.
Somatic mutations of MCL1:
Generating mutation plots.
Highly correlated genes for MCL1:
Showing top 20/101 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MCL1 | KIF13A | 0.727212 | 3 | 0 | 3 |
| MCL1 | SNX27 | 0.72606 | 3 | 0 | 3 |
| MCL1 | CNTD1 | 0.699834 | 3 | 0 | 3 |
| MCL1 | RILPL1 | 0.682704 | 3 | 0 | 3 |
| MCL1 | GABBR1 | 0.679611 | 5 | 0 | 3 |
| MCL1 | C20orf194 | 0.675941 | 3 | 0 | 3 |
| MCL1 | RNF115 | 0.66425 | 4 | 0 | 4 |
| MCL1 | OCRL | 0.652013 | 4 | 0 | 4 |
| MCL1 | IER2 | 0.648794 | 7 | 0 | 7 |
| MCL1 | SIK1 | 0.646909 | 4 | 0 | 3 |
| MCL1 | DNAJC18 | 0.646184 | 3 | 0 | 3 |
| MCL1 | APLP2 | 0.645894 | 4 | 0 | 3 |
| MCL1 | SPAG7 | 0.645001 | 4 | 0 | 3 |
| MCL1 | PITPNB | 0.643864 | 4 | 0 | 3 |
| MCL1 | PPP1R21 | 0.643533 | 4 | 0 | 3 |
| MCL1 | MIDN | 0.642078 | 5 | 0 | 5 |
| MCL1 | MFGE8 | 0.641837 | 3 | 0 | 3 |
| MCL1 | JUN | 0.636207 | 11 | 0 | 9 |
| MCL1 | JUNB | 0.632549 | 10 | 0 | 9 |
| MCL1 | TTC7B | 0.631093 | 3 | 0 | 3 |
For details and further investigation, click here