| Full name: gamma-aminobutyric acid type B receptor subunit 1 | Alias Symbol: hGB1a|GPRC3A | ||
| Type: protein-coding gene | Cytoband: 6p22.1 | ||
| Entrez ID: 2550 | HGNC ID: HGNC:4070 | Ensembl Gene: ENSG00000204681 | OMIM ID: 603540 |
| Drug and gene relationship at DGIdb | |||
GABBR1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04024 | cAMP signaling pathway | |
| hsa04727 | GABAergic synapse | |
| hsa04742 | Taste transduction | |
| hsa04915 | Estrogen signaling pathway | |
| hsa05032 | Morphine addiction |
Expression of GABBR1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GABBR1 | 2550 | 203146_s_at | -0.1113 | 0.8942 | |
| GSE20347 | GABBR1 | 2550 | 203146_s_at | 0.0914 | 0.4849 | |
| GSE23400 | GABBR1 | 2550 | 203146_s_at | -0.1356 | 0.1004 | |
| GSE26886 | GABBR1 | 2550 | 203146_s_at | -0.1355 | 0.5156 | |
| GSE29001 | GABBR1 | 2550 | 203146_s_at | 0.2592 | 0.5436 | |
| GSE38129 | GABBR1 | 2550 | 203146_s_at | -0.1571 | 0.4073 | |
| GSE45670 | GABBR1 | 2550 | 203146_s_at | -0.1321 | 0.5895 | |
| GSE53622 | GABBR1 | 2550 | 83765 | 0.2892 | 0.0225 | |
| GSE53624 | GABBR1 | 2550 | 83765 | 0.2093 | 0.0222 | |
| GSE63941 | GABBR1 | 2550 | 203146_s_at | -0.2918 | 0.2597 | |
| GSE77861 | GABBR1 | 2550 | 203146_s_at | 0.2424 | 0.4783 | |
| GSE97050 | GABBR1 | 2550 | A_23_P93302 | -0.7763 | 0.0830 | |
| SRP007169 | GABBR1 | 2550 | RNAseq | 0.8963 | 0.1664 | |
| SRP064894 | GABBR1 | 2550 | RNAseq | -0.0194 | 0.9406 | |
| SRP133303 | GABBR1 | 2550 | RNAseq | -0.2263 | 0.3640 | |
| SRP159526 | GABBR1 | 2550 | RNAseq | 0.2254 | 0.4881 | |
| SRP193095 | GABBR1 | 2550 | RNAseq | 1.1097 | 0.0000 | |
| SRP219564 | GABBR1 | 2550 | RNAseq | 0.1657 | 0.6631 | |
| TCGA | GABBR1 | 2550 | RNAseq | -0.1087 | 0.1950 |
Upregulated datasets: 1; Downregulated datasets: 0.
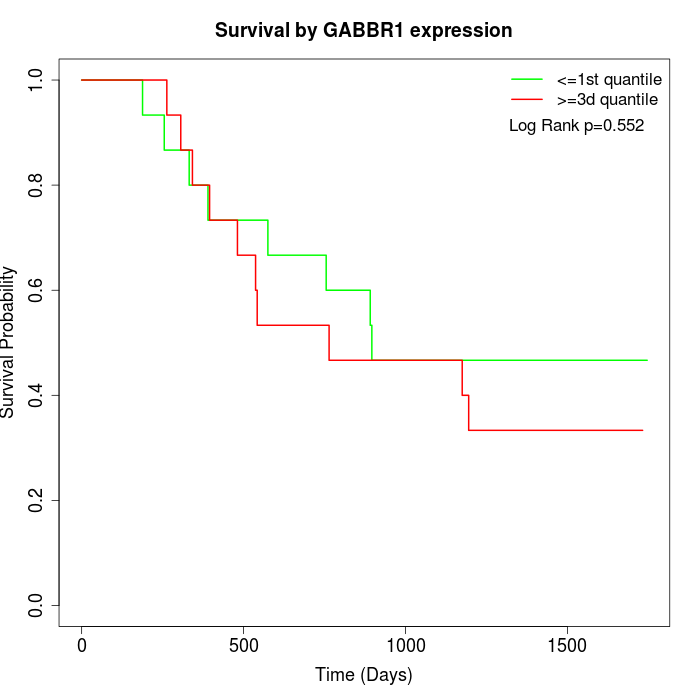
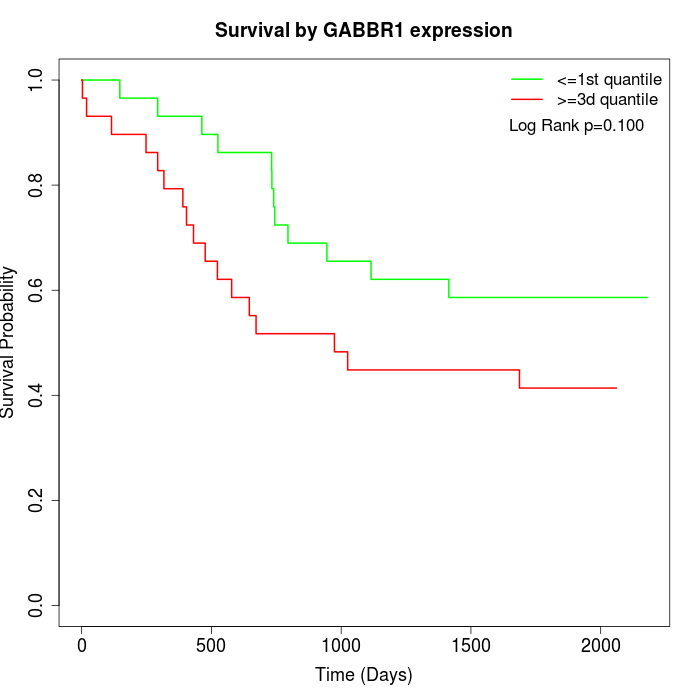
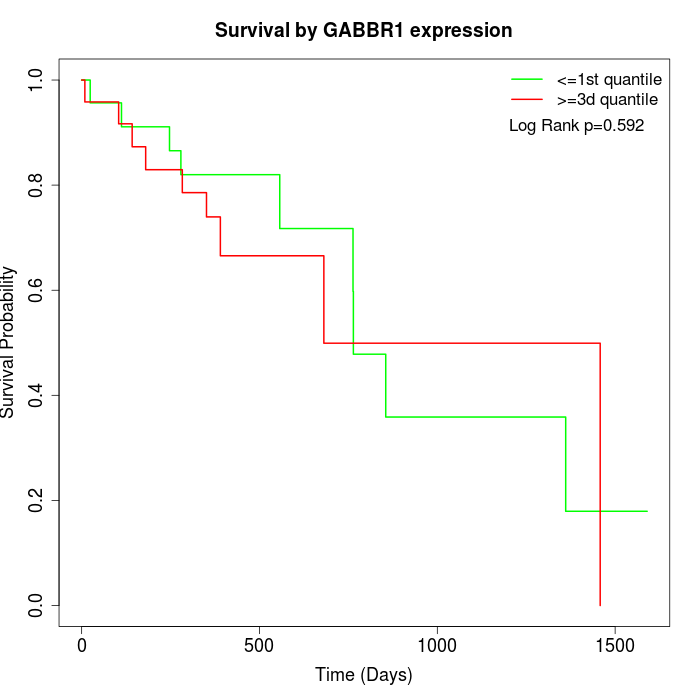
Survival by GABBR1 expression:
Note: Click image to view full size file.
Copy number change of GABBR1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GABBR1 | 2550 | 2 | 2 | 26 | |
| GSE20123 | GABBR1 | 2550 | 2 | 2 | 26 | |
| GSE43470 | GABBR1 | 2550 | 5 | 0 | 38 | |
| GSE46452 | GABBR1 | 2550 | 1 | 10 | 48 | |
| GSE47630 | GABBR1 | 2550 | 7 | 4 | 29 | |
| GSE54993 | GABBR1 | 2550 | 2 | 1 | 67 | |
| GSE54994 | GABBR1 | 2550 | 11 | 4 | 38 | |
| GSE60625 | GABBR1 | 2550 | 0 | 1 | 10 | |
| GSE74703 | GABBR1 | 2550 | 5 | 0 | 31 | |
| GSE74704 | GABBR1 | 2550 | 0 | 1 | 19 | |
| TCGA | GABBR1 | 2550 | 16 | 18 | 62 |
Total number of gains: 51; Total number of losses: 43; Total Number of normals: 394.
Somatic mutations of GABBR1:
Generating mutation plots.
Highly correlated genes for GABBR1:
Showing top 20/433 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GABBR1 | FOXC1 | 0.763569 | 3 | 0 | 3 |
| GABBR1 | PTAR1 | 0.733918 | 3 | 0 | 3 |
| GABBR1 | GTPBP1 | 0.732138 | 4 | 0 | 4 |
| GABBR1 | CCDC149 | 0.730938 | 3 | 0 | 3 |
| GABBR1 | C17orf67 | 0.730901 | 3 | 0 | 3 |
| GABBR1 | LENG8 | 0.725266 | 4 | 0 | 4 |
| GABBR1 | SLC19A2 | 0.714895 | 3 | 0 | 3 |
| GABBR1 | ZMAT1 | 0.709404 | 3 | 0 | 3 |
| GABBR1 | C20orf194 | 0.709395 | 3 | 0 | 3 |
| GABBR1 | SLITRK3 | 0.703603 | 3 | 0 | 3 |
| GABBR1 | ING1 | 0.702269 | 3 | 0 | 3 |
| GABBR1 | LONRF3 | 0.697151 | 3 | 0 | 3 |
| GABBR1 | SNX27 | 0.694382 | 5 | 0 | 4 |
| GABBR1 | SYNC | 0.692061 | 3 | 0 | 3 |
| GABBR1 | PRR12 | 0.691268 | 3 | 0 | 3 |
| GABBR1 | FXYD6 | 0.686888 | 6 | 0 | 6 |
| GABBR1 | IPO11 | 0.685027 | 3 | 0 | 3 |
| GABBR1 | ME3 | 0.683556 | 4 | 0 | 4 |
| GABBR1 | KCNJ8 | 0.680031 | 4 | 0 | 4 |
| GABBR1 | MCL1 | 0.679611 | 5 | 0 | 3 |
For details and further investigation, click here