| Full name: Jun proto-oncogene, AP-1 transcription factor subunit | Alias Symbol: c-Jun|AP-1 | ||
| Type: protein-coding gene | Cytoband: 1p32.1 | ||
| Entrez ID: 3725 | HGNC ID: HGNC:6204 | Ensembl Gene: ENSG00000177606 | OMIM ID: 165160 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
JUN involved pathways:
Expression of JUN:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | JUN | 3725 | 201464_x_at | -0.5118 | 0.4115 | |
| GSE20347 | JUN | 3725 | 201464_x_at | -0.0509 | 0.8499 | |
| GSE23400 | JUN | 3725 | 201464_x_at | -0.0850 | 0.4823 | |
| GSE26886 | JUN | 3725 | 201464_x_at | 0.9090 | 0.0034 | |
| GSE29001 | JUN | 3725 | 201464_x_at | -0.2085 | 0.4374 | |
| GSE38129 | JUN | 3725 | 201464_x_at | -0.2993 | 0.1750 | |
| GSE45670 | JUN | 3725 | 201464_x_at | -0.7149 | 0.0148 | |
| GSE53622 | JUN | 3725 | 85800 | -1.0034 | 0.0000 | |
| GSE53624 | JUN | 3725 | 85800 | -0.6650 | 0.0000 | |
| GSE63941 | JUN | 3725 | 201464_x_at | -1.1979 | 0.2028 | |
| GSE77861 | JUN | 3725 | 201464_x_at | 0.1557 | 0.7723 | |
| GSE97050 | JUN | 3725 | A_33_P3323298 | -0.9482 | 0.2172 | |
| SRP007169 | JUN | 3725 | RNAseq | -1.0880 | 0.0267 | |
| SRP008496 | JUN | 3725 | RNAseq | -0.8750 | 0.0182 | |
| SRP064894 | JUN | 3725 | RNAseq | -0.0653 | 0.7773 | |
| SRP133303 | JUN | 3725 | RNAseq | -0.3240 | 0.3449 | |
| SRP159526 | JUN | 3725 | RNAseq | 0.0494 | 0.9311 | |
| SRP193095 | JUN | 3725 | RNAseq | 0.4479 | 0.0940 | |
| SRP219564 | JUN | 3725 | RNAseq | -0.0460 | 0.9447 | |
| TCGA | JUN | 3725 | RNAseq | -0.1388 | 0.0408 |
Upregulated datasets: 0; Downregulated datasets: 2.
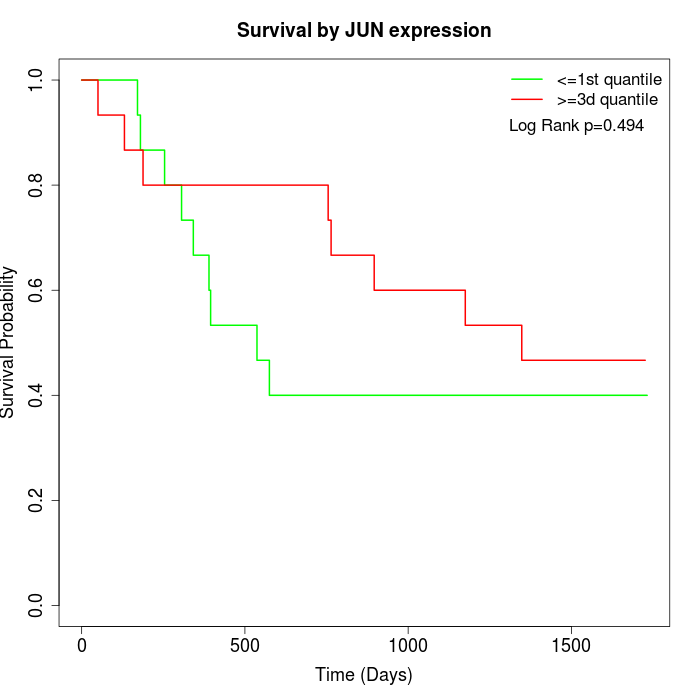
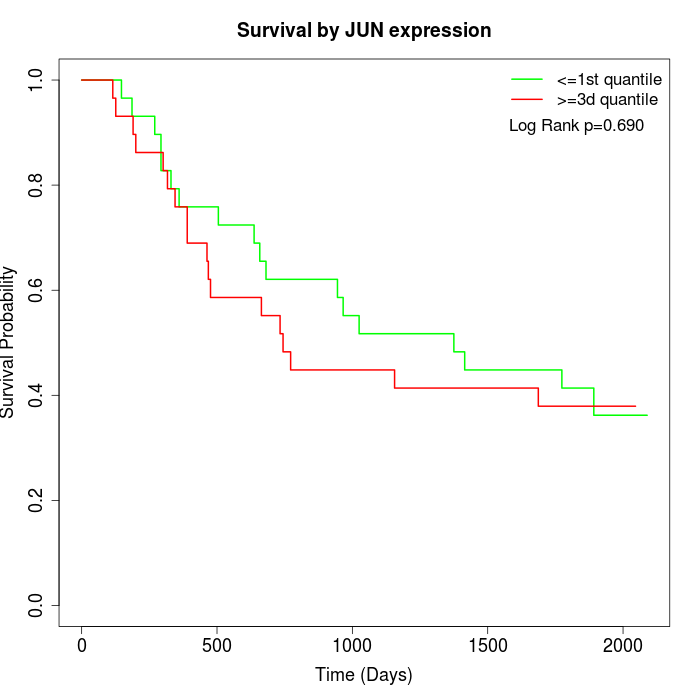
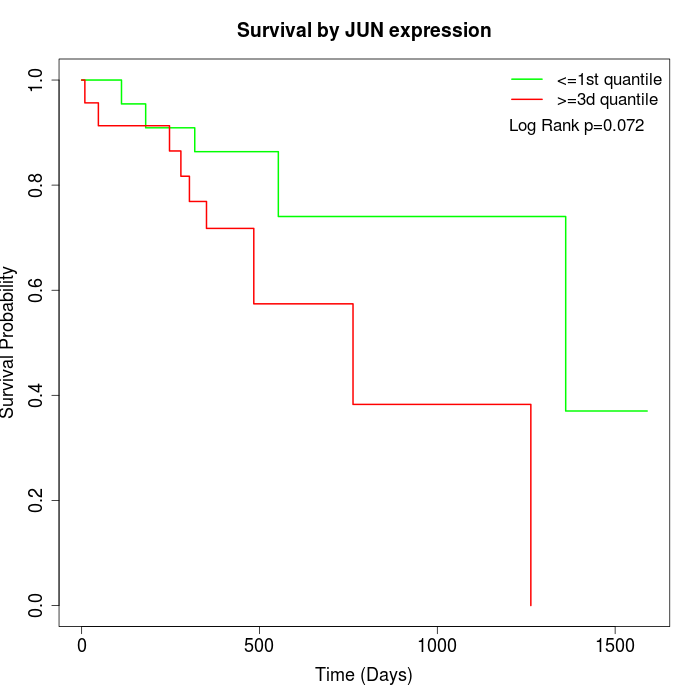
Survival by JUN expression:
Note: Click image to view full size file.
Copy number change of JUN:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | JUN | 3725 | 3 | 5 | 22 | |
| GSE20123 | JUN | 3725 | 3 | 4 | 23 | |
| GSE43470 | JUN | 3725 | 6 | 2 | 35 | |
| GSE46452 | JUN | 3725 | 2 | 1 | 56 | |
| GSE47630 | JUN | 3725 | 8 | 5 | 27 | |
| GSE54993 | JUN | 3725 | 0 | 0 | 70 | |
| GSE54994 | JUN | 3725 | 8 | 2 | 43 | |
| GSE60625 | JUN | 3725 | 0 | 0 | 11 | |
| GSE74703 | JUN | 3725 | 5 | 2 | 29 | |
| GSE74704 | JUN | 3725 | 2 | 0 | 18 | |
| TCGA | JUN | 3725 | 10 | 18 | 68 |
Total number of gains: 47; Total number of losses: 39; Total Number of normals: 402.
Somatic mutations of JUN:
Generating mutation plots.
Highly correlated genes for JUN:
Showing top 20/206 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| JUN | C9orf47 | 0.767085 | 3 | 0 | 3 |
| JUN | FOS | 0.759212 | 12 | 0 | 12 |
| JUN | KPNA1 | 0.731502 | 3 | 0 | 3 |
| JUN | FEM1C | 0.716368 | 4 | 0 | 3 |
| JUN | EGR1 | 0.71493 | 11 | 0 | 10 |
| JUN | TPBG | 0.689682 | 3 | 0 | 3 |
| JUN | WSB1 | 0.682386 | 3 | 0 | 3 |
| JUN | NRL | 0.674773 | 3 | 0 | 3 |
| JUN | NR4A1 | 0.670038 | 8 | 0 | 7 |
| JUN | LUC7L | 0.665582 | 4 | 0 | 3 |
| JUN | RRAS2 | 0.6633 | 3 | 0 | 3 |
| JUN | SLC25A25 | 0.661147 | 4 | 0 | 3 |
| JUN | C5orf24 | 0.66072 | 4 | 0 | 3 |
| JUN | CCDC40 | 0.654816 | 4 | 0 | 4 |
| JUN | FOSB | 0.65156 | 13 | 0 | 12 |
| JUN | MATN4 | 0.650221 | 3 | 0 | 3 |
| JUN | TRPC1 | 0.646188 | 4 | 0 | 3 |
| JUN | CIC | 0.644626 | 3 | 0 | 3 |
| JUN | GLI3 | 0.642918 | 3 | 0 | 3 |
| JUN | WDR27 | 0.641944 | 3 | 0 | 3 |
For details and further investigation, click here