| Full name: phosphatidylinositol transfer protein beta | Alias Symbol: VIB1B | ||
| Type: protein-coding gene | Cytoband: 22q12.1 | ||
| Entrez ID: 23760 | HGNC ID: HGNC:9002 | Ensembl Gene: ENSG00000180957 | OMIM ID: 606876 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of PITPNB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PITPNB | 23760 | 202522_at | -0.0987 | 0.6098 | |
| GSE20347 | PITPNB | 23760 | 202522_at | 0.0170 | 0.9011 | |
| GSE23400 | PITPNB | 23760 | 202522_at | 0.1028 | 0.0527 | |
| GSE26886 | PITPNB | 23760 | 202522_at | 0.1790 | 0.1650 | |
| GSE29001 | PITPNB | 23760 | 202522_at | 0.2357 | 0.1196 | |
| GSE38129 | PITPNB | 23760 | 202522_at | -0.0797 | 0.4412 | |
| GSE45670 | PITPNB | 23760 | 202522_at | -0.0437 | 0.8252 | |
| GSE53622 | PITPNB | 23760 | 3609 | -0.2022 | 0.0003 | |
| GSE53624 | PITPNB | 23760 | 3609 | 0.1859 | 0.0004 | |
| GSE63941 | PITPNB | 23760 | 202522_at | 0.5499 | 0.0205 | |
| GSE77861 | PITPNB | 23760 | 202522_at | 0.1507 | 0.4773 | |
| GSE97050 | PITPNB | 23760 | A_24_P305678 | -0.0939 | 0.6481 | |
| SRP007169 | PITPNB | 23760 | RNAseq | 1.7085 | 0.0003 | |
| SRP008496 | PITPNB | 23760 | RNAseq | 1.2411 | 0.0003 | |
| SRP064894 | PITPNB | 23760 | RNAseq | 0.1194 | 0.4193 | |
| SRP133303 | PITPNB | 23760 | RNAseq | 0.1966 | 0.1850 | |
| SRP159526 | PITPNB | 23760 | RNAseq | -0.1311 | 0.5772 | |
| SRP193095 | PITPNB | 23760 | RNAseq | 0.0991 | 0.3173 | |
| SRP219564 | PITPNB | 23760 | RNAseq | 0.1003 | 0.7773 | |
| TCGA | PITPNB | 23760 | RNAseq | 0.0648 | 0.1464 |
Upregulated datasets: 2; Downregulated datasets: 0.
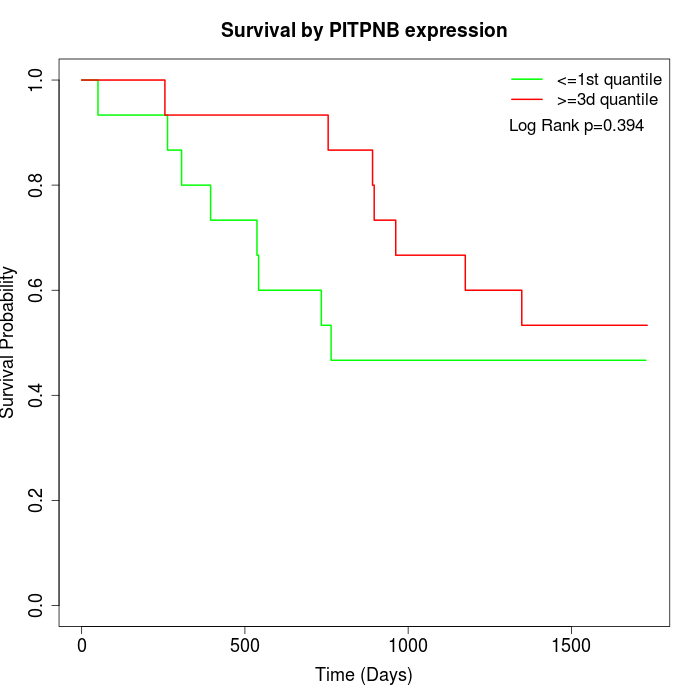
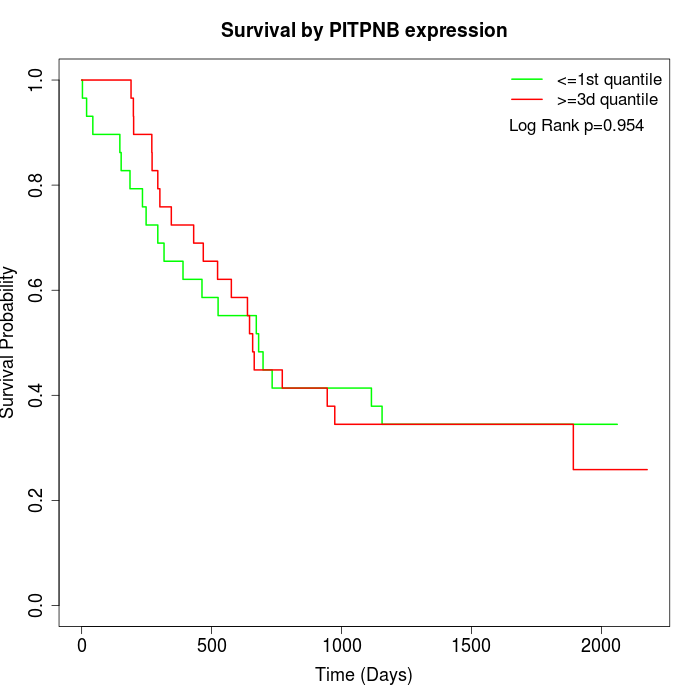
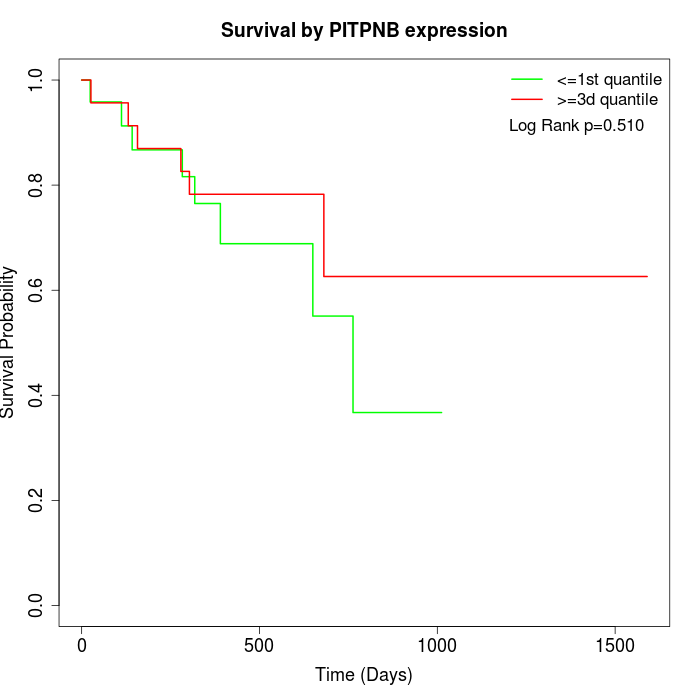
Survival by PITPNB expression:
Note: Click image to view full size file.
Copy number change of PITPNB:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PITPNB | 23760 | 2 | 5 | 23 | |
| GSE20123 | PITPNB | 23760 | 2 | 4 | 24 | |
| GSE43470 | PITPNB | 23760 | 3 | 6 | 34 | |
| GSE46452 | PITPNB | 23760 | 31 | 1 | 27 | |
| GSE47630 | PITPNB | 23760 | 8 | 5 | 27 | |
| GSE54993 | PITPNB | 23760 | 3 | 6 | 61 | |
| GSE54994 | PITPNB | 23760 | 11 | 8 | 34 | |
| GSE60625 | PITPNB | 23760 | 5 | 0 | 6 | |
| GSE74703 | PITPNB | 23760 | 3 | 5 | 28 | |
| GSE74704 | PITPNB | 23760 | 0 | 1 | 19 | |
| TCGA | PITPNB | 23760 | 28 | 16 | 52 |
Total number of gains: 96; Total number of losses: 57; Total Number of normals: 335.
Somatic mutations of PITPNB:
Generating mutation plots.
Highly correlated genes for PITPNB:
Showing top 20/129 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PITPNB | CNTD1 | 0.760089 | 3 | 0 | 3 |
| PITPNB | C5orf24 | 0.743974 | 3 | 0 | 3 |
| PITPNB | BEX5 | 0.738787 | 3 | 0 | 3 |
| PITPNB | ZSCAN12 | 0.719302 | 3 | 0 | 3 |
| PITPNB | SERINC1 | 0.711095 | 3 | 0 | 3 |
| PITPNB | COCH | 0.711008 | 3 | 0 | 3 |
| PITPNB | TTF1 | 0.710152 | 3 | 0 | 3 |
| PITPNB | PPIE | 0.706055 | 3 | 0 | 3 |
| PITPNB | IRF2BPL | 0.705417 | 3 | 0 | 3 |
| PITPNB | SLMAP | 0.70491 | 3 | 0 | 3 |
| PITPNB | PRKRA | 0.701151 | 4 | 0 | 3 |
| PITPNB | VPS72 | 0.700828 | 3 | 0 | 3 |
| PITPNB | RABGAP1 | 0.697651 | 3 | 0 | 3 |
| PITPNB | CDADC1 | 0.696721 | 4 | 0 | 3 |
| PITPNB | BRD4 | 0.694769 | 3 | 0 | 3 |
| PITPNB | ZNF814 | 0.691789 | 3 | 0 | 3 |
| PITPNB | NDUFS6 | 0.685748 | 3 | 0 | 3 |
| PITPNB | DIAPH2 | 0.682623 | 3 | 0 | 3 |
| PITPNB | EIF3G | 0.677349 | 3 | 0 | 3 |
| PITPNB | ANO6 | 0.676695 | 4 | 0 | 3 |
For details and further investigation, click here