| Full name: JunB proto-oncogene, AP-1 transcription factor subunit | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 19p13.13 | ||
| Entrez ID: 3726 | HGNC ID: HGNC:6205 | Ensembl Gene: ENSG00000171223 | OMIM ID: 165161 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
JUNB involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04380 | Osteoclast differentiation |
Expression of JUNB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | JUNB | 3726 | 201473_at | -0.8907 | 0.2792 | |
| GSE20347 | JUNB | 3726 | 201473_at | -0.5467 | 0.0169 | |
| GSE23400 | JUNB | 3726 | 201473_at | -0.6544 | 0.0000 | |
| GSE26886 | JUNB | 3726 | 201473_at | -0.3530 | 0.1428 | |
| GSE29001 | JUNB | 3726 | 201473_at | -0.6938 | 0.0213 | |
| GSE38129 | JUNB | 3726 | 201473_at | -0.6601 | 0.0036 | |
| GSE45670 | JUNB | 3726 | 201473_at | -0.2100 | 0.2323 | |
| GSE53622 | JUNB | 3726 | 10015 | -0.4132 | 0.0004 | |
| GSE53624 | JUNB | 3726 | 10015 | -0.2878 | 0.0002 | |
| GSE63941 | JUNB | 3726 | 201473_at | 1.2625 | 0.0722 | |
| GSE77861 | JUNB | 3726 | 201473_at | 0.1685 | 0.5384 | |
| GSE97050 | JUNB | 3726 | A_24_P241815 | -0.1375 | 0.7900 | |
| SRP007169 | JUNB | 3726 | RNAseq | -2.8494 | 0.0000 | |
| SRP008496 | JUNB | 3726 | RNAseq | -2.1900 | 0.0000 | |
| SRP064894 | JUNB | 3726 | RNAseq | -0.7033 | 0.0130 | |
| SRP133303 | JUNB | 3726 | RNAseq | -0.4781 | 0.0662 | |
| SRP159526 | JUNB | 3726 | RNAseq | -0.6514 | 0.1197 | |
| SRP193095 | JUNB | 3726 | RNAseq | -0.1902 | 0.3775 | |
| SRP219564 | JUNB | 3726 | RNAseq | -0.8327 | 0.0301 | |
| TCGA | JUNB | 3726 | RNAseq | 0.0289 | 0.6736 |
Upregulated datasets: 0; Downregulated datasets: 2.
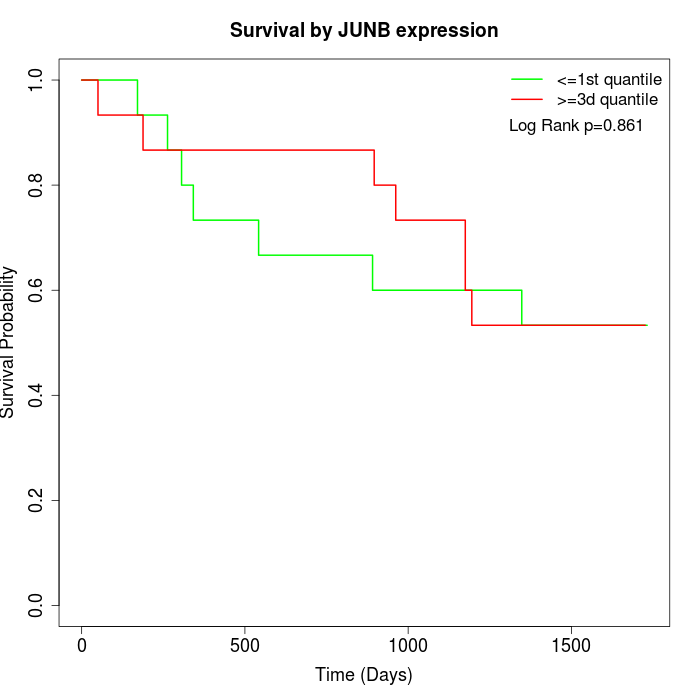
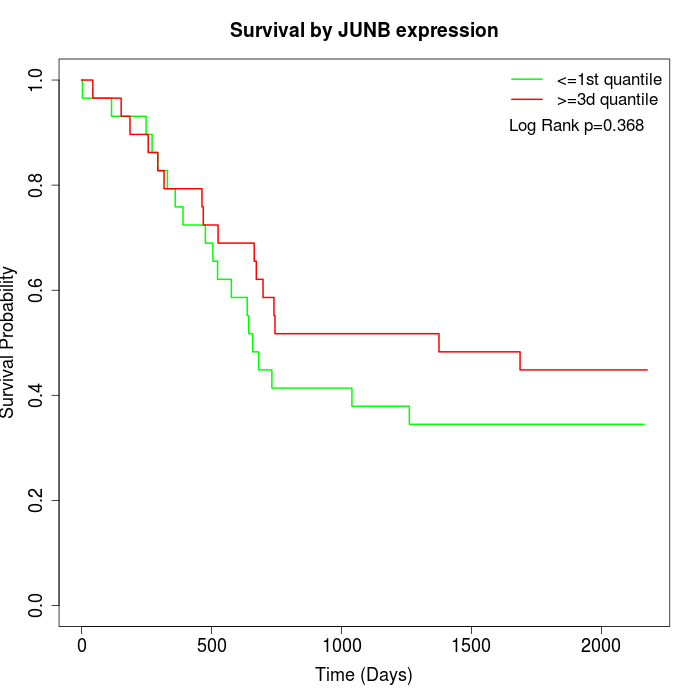
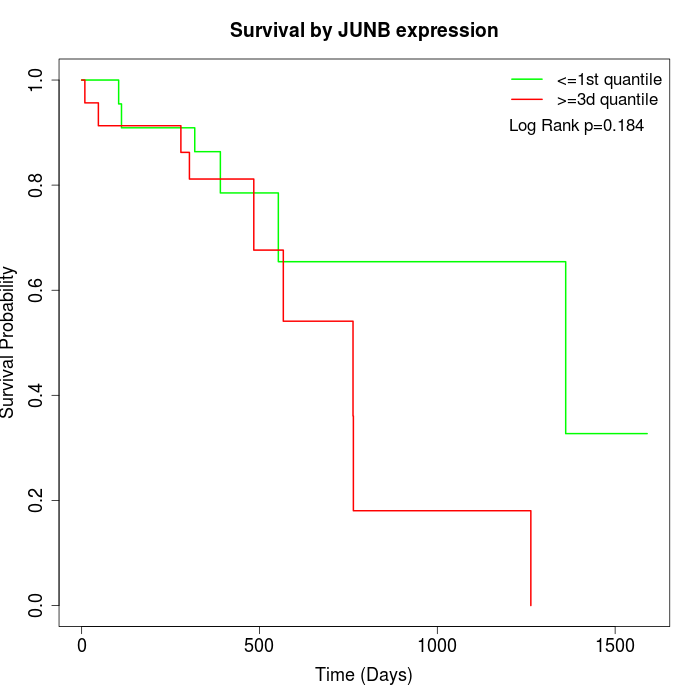
Survival by JUNB expression:
Note: Click image to view full size file.
Copy number change of JUNB:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | JUNB | 3726 | 4 | 2 | 24 | |
| GSE20123 | JUNB | 3726 | 3 | 1 | 26 | |
| GSE43470 | JUNB | 3726 | 2 | 6 | 35 | |
| GSE46452 | JUNB | 3726 | 47 | 1 | 11 | |
| GSE47630 | JUNB | 3726 | 4 | 7 | 29 | |
| GSE54993 | JUNB | 3726 | 16 | 3 | 51 | |
| GSE54994 | JUNB | 3726 | 6 | 13 | 34 | |
| GSE60625 | JUNB | 3726 | 9 | 0 | 2 | |
| GSE74703 | JUNB | 3726 | 2 | 4 | 30 | |
| GSE74704 | JUNB | 3726 | 0 | 1 | 19 | |
| TCGA | JUNB | 3726 | 16 | 13 | 67 |
Total number of gains: 109; Total number of losses: 51; Total Number of normals: 328.
Somatic mutations of JUNB:
Generating mutation plots.
Highly correlated genes for JUNB:
Showing top 20/139 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| JUNB | ADAMTS10 | 0.755595 | 3 | 0 | 3 |
| JUNB | IER2 | 0.745859 | 11 | 0 | 10 |
| JUNB | DUSP1 | 0.731374 | 11 | 0 | 11 |
| JUNB | GPR75 | 0.714066 | 3 | 0 | 3 |
| JUNB | ZFP36 | 0.687232 | 11 | 0 | 9 |
| JUNB | FIBCD1 | 0.683077 | 3 | 0 | 3 |
| JUNB | SIK1 | 0.681433 | 5 | 0 | 5 |
| JUNB | FOS | 0.675144 | 12 | 0 | 10 |
| JUNB | WDR93 | 0.669439 | 3 | 0 | 3 |
| JUNB | EGR1 | 0.652571 | 11 | 0 | 11 |
| JUNB | CSRNP1 | 0.646931 | 6 | 0 | 6 |
| JUNB | PLK3 | 0.641187 | 8 | 0 | 7 |
| JUNB | SLC25A25 | 0.636463 | 5 | 0 | 4 |
| JUNB | NR4A1 | 0.635408 | 6 | 0 | 5 |
| JUNB | RRAGA | 0.633203 | 3 | 0 | 3 |
| JUNB | MCL1 | 0.632549 | 10 | 0 | 9 |
| JUNB | MAN2C1 | 0.631548 | 4 | 0 | 4 |
| JUNB | FOSB | 0.630397 | 11 | 0 | 9 |
| JUNB | CCND2 | 0.629142 | 4 | 0 | 3 |
| JUNB | NLRP12 | 0.623572 | 4 | 0 | 3 |
For details and further investigation, click here