| Full name: NCK adaptor protein 2 | Alias Symbol: NCKbeta | ||
| Type: protein-coding gene | Cytoband: 2q12.2 | ||
| Entrez ID: 8440 | HGNC ID: HGNC:7665 | Ensembl Gene: ENSG00000071051 | OMIM ID: 604930 |
| Drug and gene relationship at DGIdb | |||
NCK2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04012 | ErbB signaling pathway | |
| hsa04360 | Axon guidance | |
| hsa04660 | T cell receptor signaling pathway | |
| hsa05130 | Pathogenic Escherichia coli infection |
Expression of NCK2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NCK2 | 8440 | 203315_at | -0.3898 | 0.1346 | |
| GSE20347 | NCK2 | 8440 | 203315_at | -0.8149 | 0.0000 | |
| GSE23400 | NCK2 | 8440 | 203315_at | -0.4704 | 0.0000 | |
| GSE26886 | NCK2 | 8440 | 203315_at | -0.7885 | 0.0000 | |
| GSE29001 | NCK2 | 8440 | 203315_at | -0.6748 | 0.0155 | |
| GSE38129 | NCK2 | 8440 | 203315_at | -0.5762 | 0.0001 | |
| GSE45670 | NCK2 | 8440 | 203315_at | -0.2874 | 0.0584 | |
| GSE53622 | NCK2 | 8440 | 13179 | -0.6688 | 0.0000 | |
| GSE53624 | NCK2 | 8440 | 13179 | -0.8722 | 0.0000 | |
| GSE63941 | NCK2 | 8440 | 203315_at | -0.6927 | 0.2518 | |
| GSE77861 | NCK2 | 8440 | 203315_at | -0.7868 | 0.0281 | |
| GSE97050 | NCK2 | 8440 | A_33_P3268564 | -0.1456 | 0.4467 | |
| SRP007169 | NCK2 | 8440 | RNAseq | -1.9258 | 0.0000 | |
| SRP008496 | NCK2 | 8440 | RNAseq | -1.6760 | 0.0000 | |
| SRP064894 | NCK2 | 8440 | RNAseq | -1.2709 | 0.0000 | |
| SRP133303 | NCK2 | 8440 | RNAseq | -0.6153 | 0.0004 | |
| SRP159526 | NCK2 | 8440 | RNAseq | -0.6741 | 0.0423 | |
| SRP193095 | NCK2 | 8440 | RNAseq | -1.0943 | 0.0000 | |
| SRP219564 | NCK2 | 8440 | RNAseq | -1.4923 | 0.0001 | |
| TCGA | NCK2 | 8440 | RNAseq | 0.0108 | 0.8320 |
Upregulated datasets: 0; Downregulated datasets: 5.
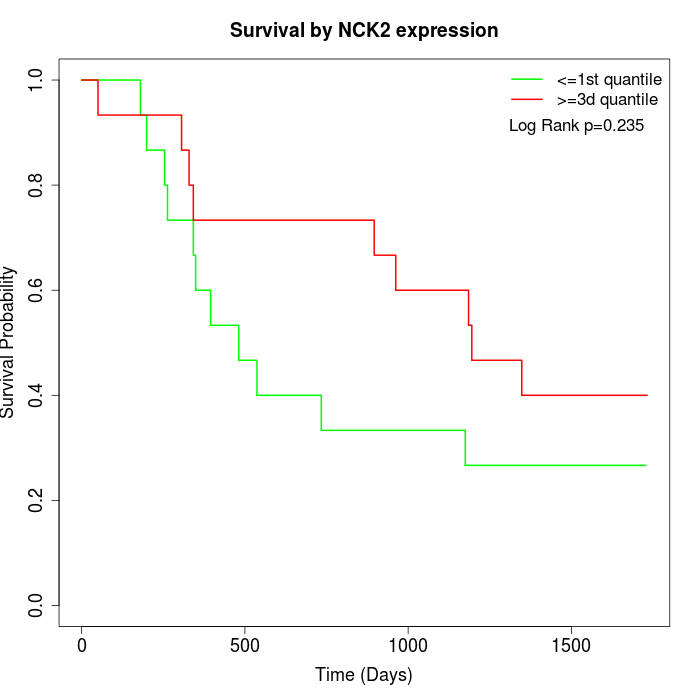
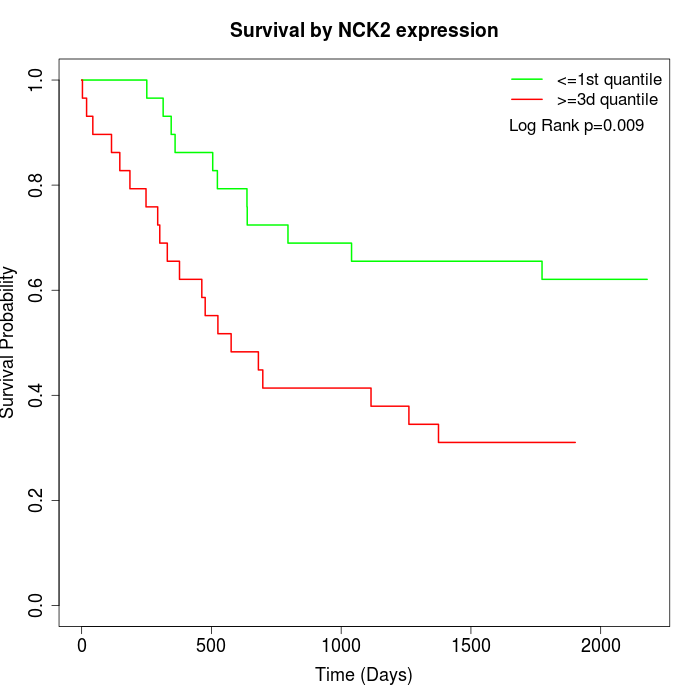
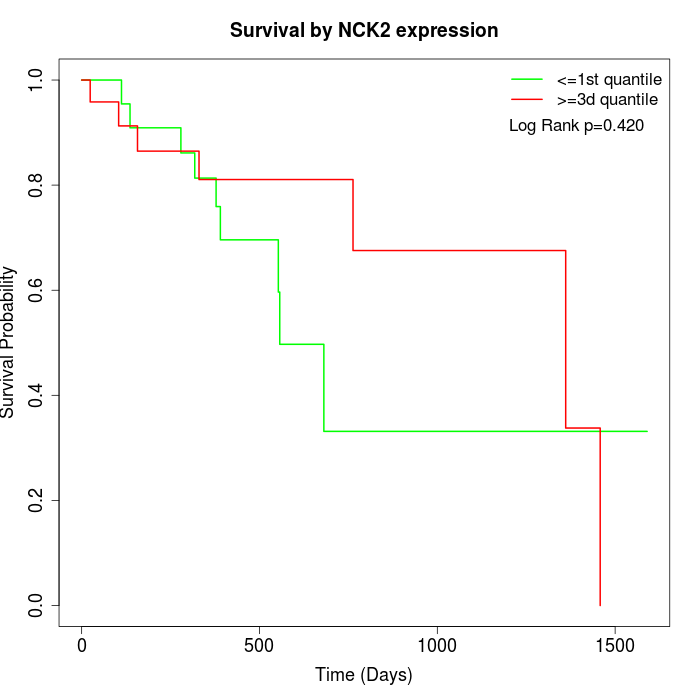
Survival by NCK2 expression:
Note: Click image to view full size file.
Copy number change of NCK2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NCK2 | 8440 | 3 | 3 | 24 | |
| GSE20123 | NCK2 | 8440 | 3 | 3 | 24 | |
| GSE43470 | NCK2 | 8440 | 3 | 1 | 39 | |
| GSE46452 | NCK2 | 8440 | 1 | 4 | 54 | |
| GSE47630 | NCK2 | 8440 | 7 | 0 | 33 | |
| GSE54993 | NCK2 | 8440 | 0 | 6 | 64 | |
| GSE54994 | NCK2 | 8440 | 10 | 0 | 43 | |
| GSE60625 | NCK2 | 8440 | 0 | 3 | 8 | |
| GSE74703 | NCK2 | 8440 | 3 | 1 | 32 | |
| GSE74704 | NCK2 | 8440 | 2 | 3 | 15 | |
| TCGA | NCK2 | 8440 | 35 | 6 | 55 |
Total number of gains: 67; Total number of losses: 30; Total Number of normals: 391.
Somatic mutations of NCK2:
Generating mutation plots.
Highly correlated genes for NCK2:
Showing top 20/1534 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NCK2 | MMEL1 | 0.803748 | 4 | 0 | 4 |
| NCK2 | ZFAND2B | 0.790523 | 7 | 0 | 7 |
| NCK2 | SFTA2 | 0.784499 | 4 | 0 | 4 |
| NCK2 | SMIM5 | 0.784176 | 6 | 0 | 6 |
| NCK2 | SNORA68 | 0.78263 | 4 | 0 | 4 |
| NCK2 | ATP6V1G1 | 0.782628 | 3 | 0 | 3 |
| NCK2 | ARPC1A | 0.768801 | 3 | 0 | 3 |
| NCK2 | PLEKHA7 | 0.759521 | 7 | 0 | 7 |
| NCK2 | ELMO2 | 0.756228 | 10 | 0 | 10 |
| NCK2 | YPEL3 | 0.754701 | 7 | 0 | 7 |
| NCK2 | VSIG10L | 0.751483 | 6 | 0 | 6 |
| NCK2 | TBC1D23 | 0.748779 | 3 | 0 | 3 |
| NCK2 | FAM214A | 0.746624 | 6 | 0 | 6 |
| NCK2 | ARHGAP27 | 0.745461 | 7 | 0 | 7 |
| NCK2 | KRT78 | 0.744124 | 6 | 0 | 6 |
| NCK2 | UBL3 | 0.740772 | 10 | 0 | 10 |
| NCK2 | CYSRT1 | 0.740594 | 6 | 0 | 6 |
| NCK2 | GGA3 | 0.737977 | 4 | 0 | 4 |
| NCK2 | CCNG2 | 0.737083 | 11 | 0 | 11 |
| NCK2 | RMND5B | 0.735499 | 11 | 0 | 11 |
For details and further investigation, click here