| Full name: engulfment and cell motility 2 | Alias Symbol: CED12|ELMO-2|CED-12|KIAA1834|FLJ11656 | ||
| Type: protein-coding gene | Cytoband: 20q13.12 | ||
| Entrez ID: 63916 | HGNC ID: HGNC:17233 | Ensembl Gene: ENSG00000062598 | OMIM ID: 606421 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ELMO2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05100 | Bacterial invasion of epithelial cells | |
| hsa05131 | Shigellosis |
Expression of ELMO2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ELMO2 | 63916 | 55692_at | -0.2234 | 0.7338 | |
| GSE20347 | ELMO2 | 63916 | 55692_at | -1.2276 | 0.0000 | |
| GSE23400 | ELMO2 | 63916 | 221528_s_at | -0.6957 | 0.0000 | |
| GSE26886 | ELMO2 | 63916 | 55692_at | -1.5966 | 0.0000 | |
| GSE29001 | ELMO2 | 63916 | 55692_at | -0.9641 | 0.0011 | |
| GSE38129 | ELMO2 | 63916 | 55692_at | -0.8636 | 0.0000 | |
| GSE45670 | ELMO2 | 63916 | 55692_at | -0.5249 | 0.0186 | |
| GSE53622 | ELMO2 | 63916 | 12160 | -0.7156 | 0.0000 | |
| GSE53624 | ELMO2 | 63916 | 12160 | -0.9636 | 0.0000 | |
| GSE63941 | ELMO2 | 63916 | 55692_at | -1.0681 | 0.0415 | |
| GSE77861 | ELMO2 | 63916 | 55692_at | -1.1424 | 0.0006 | |
| GSE97050 | ELMO2 | 63916 | A_23_P210538 | -0.4390 | 0.1902 | |
| SRP007169 | ELMO2 | 63916 | RNAseq | -1.6726 | 0.0000 | |
| SRP008496 | ELMO2 | 63916 | RNAseq | -1.5417 | 0.0000 | |
| SRP064894 | ELMO2 | 63916 | RNAseq | -0.9993 | 0.0000 | |
| SRP133303 | ELMO2 | 63916 | RNAseq | -0.7629 | 0.0000 | |
| SRP159526 | ELMO2 | 63916 | RNAseq | -0.8158 | 0.0088 | |
| SRP193095 | ELMO2 | 63916 | RNAseq | -0.8854 | 0.0000 | |
| SRP219564 | ELMO2 | 63916 | RNAseq | -0.7187 | 0.0130 | |
| TCGA | ELMO2 | 63916 | RNAseq | -0.0234 | 0.6283 |
Upregulated datasets: 0; Downregulated datasets: 6.
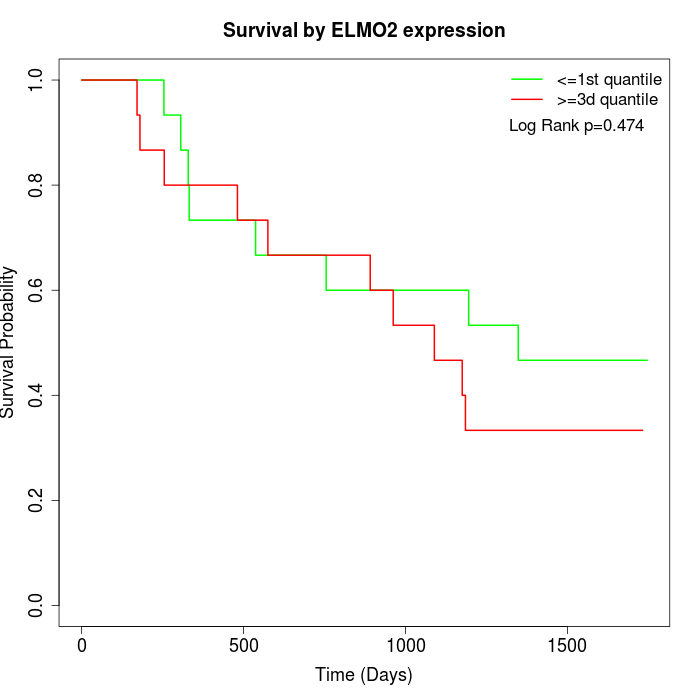
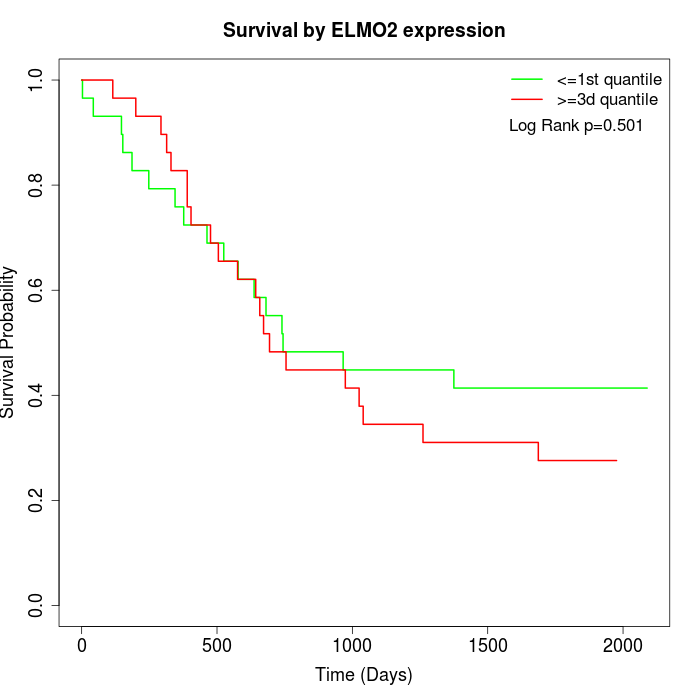
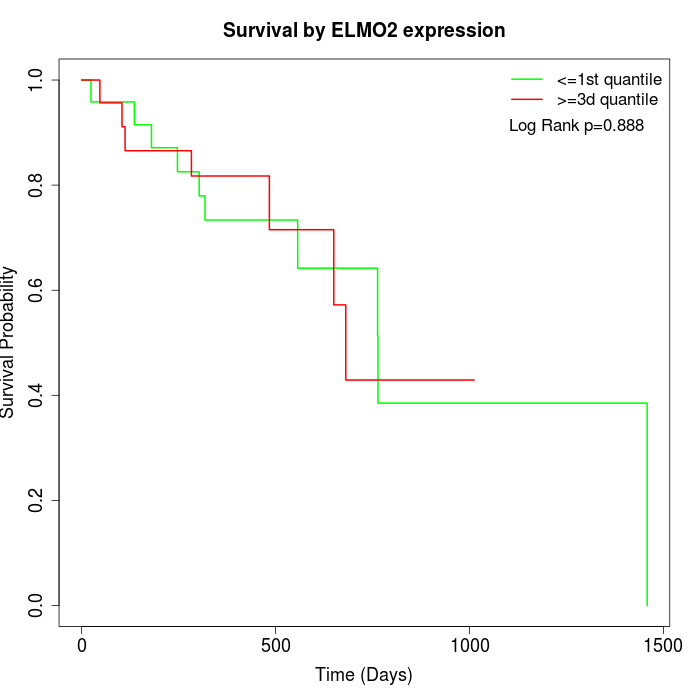
Survival by ELMO2 expression:
Note: Click image to view full size file.
Copy number change of ELMO2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ELMO2 | 63916 | 14 | 1 | 15 | |
| GSE20123 | ELMO2 | 63916 | 14 | 1 | 15 | |
| GSE43470 | ELMO2 | 63916 | 12 | 1 | 30 | |
| GSE46452 | ELMO2 | 63916 | 29 | 0 | 30 | |
| GSE47630 | ELMO2 | 63916 | 24 | 1 | 15 | |
| GSE54993 | ELMO2 | 63916 | 0 | 17 | 53 | |
| GSE54994 | ELMO2 | 63916 | 25 | 0 | 28 | |
| GSE60625 | ELMO2 | 63916 | 0 | 0 | 11 | |
| GSE74703 | ELMO2 | 63916 | 10 | 1 | 25 | |
| GSE74704 | ELMO2 | 63916 | 10 | 0 | 10 | |
| TCGA | ELMO2 | 63916 | 46 | 3 | 47 |
Total number of gains: 184; Total number of losses: 25; Total Number of normals: 279.
Somatic mutations of ELMO2:
Generating mutation plots.
Highly correlated genes for ELMO2:
Showing top 20/1657 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ELMO2 | RBM20 | 0.819444 | 4 | 0 | 4 |
| ELMO2 | UBL3 | 0.819074 | 10 | 0 | 10 |
| ELMO2 | RANBP9 | 0.818608 | 11 | 0 | 11 |
| ELMO2 | SORT1 | 0.805937 | 10 | 0 | 10 |
| ELMO2 | GYS2 | 0.80287 | 10 | 0 | 10 |
| ELMO2 | C18orf25 | 0.798919 | 11 | 0 | 11 |
| ELMO2 | ZNRF1 | 0.798769 | 4 | 0 | 4 |
| ELMO2 | ETFDH | 0.797587 | 10 | 0 | 10 |
| ELMO2 | SFTA2 | 0.796879 | 4 | 0 | 4 |
| ELMO2 | ITCH | 0.79624 | 10 | 0 | 10 |
| ELMO2 | ZNF426 | 0.794542 | 11 | 0 | 11 |
| ELMO2 | COG6 | 0.79294 | 4 | 0 | 4 |
| ELMO2 | SNX24 | 0.788933 | 9 | 0 | 9 |
| ELMO2 | CCNYL1 | 0.787477 | 5 | 0 | 5 |
| ELMO2 | PDCD6IP | 0.78706 | 11 | 0 | 11 |
| ELMO2 | CCNG2 | 0.78595 | 11 | 0 | 11 |
| ELMO2 | GPD1L | 0.783331 | 10 | 0 | 10 |
| ELMO2 | LRP10 | 0.781754 | 10 | 0 | 10 |
| ELMO2 | ACAA1 | 0.781503 | 10 | 0 | 10 |
| ELMO2 | FCHO2 | 0.781448 | 7 | 0 | 6 |
For details and further investigation, click here