| Full name: ATPase H+ transporting V1 subunit G1 | Alias Symbol: ATP6GL|Vma10|ATP6G|DKFZp547P234 | ||
| Type: protein-coding gene | Cytoband: 9q32 | ||
| Entrez ID: 9550 | HGNC ID: HGNC:864 | Ensembl Gene: ENSG00000136888 | OMIM ID: 607296 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ATP6V1G1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04150 | mTOR signaling pathway | |
| hsa05110 | Vibrio cholerae infection | |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection |
Expression of ATP6V1G1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATP6V1G1 | 9550 | 208737_at | -0.2630 | 0.3794 | |
| GSE20347 | ATP6V1G1 | 9550 | 208737_at | -0.3035 | 0.0216 | |
| GSE23400 | ATP6V1G1 | 9550 | 208737_at | -0.0831 | 0.3372 | |
| GSE26886 | ATP6V1G1 | 9550 | 208737_at | -0.0115 | 0.9636 | |
| GSE29001 | ATP6V1G1 | 9550 | 208737_at | -0.0978 | 0.7767 | |
| GSE38129 | ATP6V1G1 | 9550 | 208737_at | -0.1275 | 0.4838 | |
| GSE45670 | ATP6V1G1 | 9550 | 208737_at | -0.0058 | 0.9780 | |
| GSE63941 | ATP6V1G1 | 9550 | 208737_at | -0.1571 | 0.6816 | |
| GSE77861 | ATP6V1G1 | 9550 | 208737_at | -0.0943 | 0.7068 | |
| GSE97050 | ATP6V1G1 | 9550 | A_23_P20882 | -0.3685 | 0.3418 | |
| SRP007169 | ATP6V1G1 | 9550 | RNAseq | -1.3190 | 0.0068 | |
| SRP008496 | ATP6V1G1 | 9550 | RNAseq | -1.0968 | 0.0001 | |
| SRP064894 | ATP6V1G1 | 9550 | RNAseq | -0.2040 | 0.2147 | |
| SRP133303 | ATP6V1G1 | 9550 | RNAseq | -0.0586 | 0.7845 | |
| SRP159526 | ATP6V1G1 | 9550 | RNAseq | -0.4746 | 0.0756 | |
| SRP193095 | ATP6V1G1 | 9550 | RNAseq | -0.6229 | 0.0002 | |
| SRP219564 | ATP6V1G1 | 9550 | RNAseq | -0.2854 | 0.4016 | |
| TCGA | ATP6V1G1 | 9550 | RNAseq | 0.0181 | 0.6989 |
Upregulated datasets: 0; Downregulated datasets: 2.
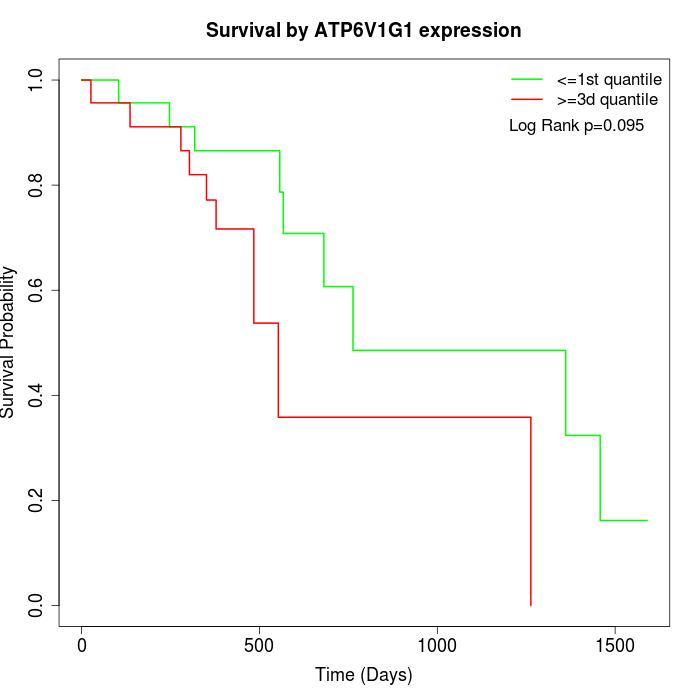
Survival by ATP6V1G1 expression:
Note: Click image to view full size file.
Copy number change of ATP6V1G1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATP6V1G1 | 9550 | 4 | 10 | 16 | |
| GSE20123 | ATP6V1G1 | 9550 | 4 | 10 | 16 | |
| GSE43470 | ATP6V1G1 | 9550 | 4 | 3 | 36 | |
| GSE46452 | ATP6V1G1 | 9550 | 6 | 13 | 40 | |
| GSE47630 | ATP6V1G1 | 9550 | 3 | 16 | 21 | |
| GSE54993 | ATP6V1G1 | 9550 | 3 | 3 | 64 | |
| GSE54994 | ATP6V1G1 | 9550 | 9 | 10 | 34 | |
| GSE60625 | ATP6V1G1 | 9550 | 0 | 0 | 11 | |
| GSE74703 | ATP6V1G1 | 9550 | 4 | 3 | 29 | |
| GSE74704 | ATP6V1G1 | 9550 | 2 | 7 | 11 | |
| TCGA | ATP6V1G1 | 9550 | 27 | 21 | 48 |
Total number of gains: 66; Total number of losses: 96; Total Number of normals: 326.
Somatic mutations of ATP6V1G1:
Generating mutation plots.
Highly correlated genes for ATP6V1G1:
Showing top 20/491 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATP6V1G1 | IMPACT | 0.783948 | 3 | 0 | 3 |
| ATP6V1G1 | NCK2 | 0.782628 | 3 | 0 | 3 |
| ATP6V1G1 | TGDS | 0.781605 | 3 | 0 | 3 |
| ATP6V1G1 | CCNYL1 | 0.780338 | 3 | 0 | 3 |
| ATP6V1G1 | ANO10 | 0.77782 | 3 | 0 | 3 |
| ATP6V1G1 | KLF3 | 0.752504 | 3 | 0 | 3 |
| ATP6V1G1 | AFTPH | 0.750999 | 3 | 0 | 3 |
| ATP6V1G1 | C2orf76 | 0.750165 | 3 | 0 | 3 |
| ATP6V1G1 | UBC | 0.746651 | 3 | 0 | 3 |
| ATP6V1G1 | FRMD6 | 0.742666 | 3 | 0 | 3 |
| ATP6V1G1 | RABGAP1L | 0.730911 | 3 | 0 | 3 |
| ATP6V1G1 | ZNF710 | 0.726739 | 3 | 0 | 3 |
| ATP6V1G1 | SCYL2 | 0.726485 | 3 | 0 | 3 |
| ATP6V1G1 | MDFIC | 0.725806 | 3 | 0 | 3 |
| ATP6V1G1 | CNOT1 | 0.724897 | 3 | 0 | 3 |
| ATP6V1G1 | YAF2 | 0.723849 | 3 | 0 | 3 |
| ATP6V1G1 | RAB2A | 0.719748 | 4 | 0 | 3 |
| ATP6V1G1 | MKNK2 | 0.71646 | 3 | 0 | 3 |
| ATP6V1G1 | MSH3 | 0.716141 | 3 | 0 | 3 |
| ATP6V1G1 | MRRF | 0.716012 | 4 | 0 | 3 |
For details and further investigation, click here