| Full name: transthyretin | Alias Symbol: HsT2651|CTS | ||
| Type: protein-coding gene | Cytoband: 18q12.1 | ||
| Entrez ID: 7276 | HGNC ID: HGNC:12405 | Ensembl Gene: ENSG00000118271 | OMIM ID: 176300 |
| Drug and gene relationship at DGIdb | |||
Expression of TTR:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TTR | 7276 | 209660_at | -0.0548 | 0.8980 | |
| GSE20347 | TTR | 7276 | 209660_at | 0.0158 | 0.8885 | |
| GSE23400 | TTR | 7276 | 209660_at | -0.1406 | 0.0002 | |
| GSE26886 | TTR | 7276 | 209660_at | -0.2872 | 0.0332 | |
| GSE29001 | TTR | 7276 | 209660_at | -0.0964 | 0.4833 | |
| GSE38129 | TTR | 7276 | 209660_at | -0.3413 | 0.0801 | |
| GSE45670 | TTR | 7276 | 209660_at | 0.0395 | 0.8079 | |
| GSE53622 | TTR | 7276 | 34443 | -1.2695 | 0.0000 | |
| GSE53624 | TTR | 7276 | 34443 | -2.1686 | 0.0000 | |
| GSE63941 | TTR | 7276 | 209660_at | 0.0468 | 0.7671 | |
| GSE77861 | TTR | 7276 | 209660_at | -0.2150 | 0.0497 | |
| TCGA | TTR | 7276 | RNAseq | -5.6415 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 3.
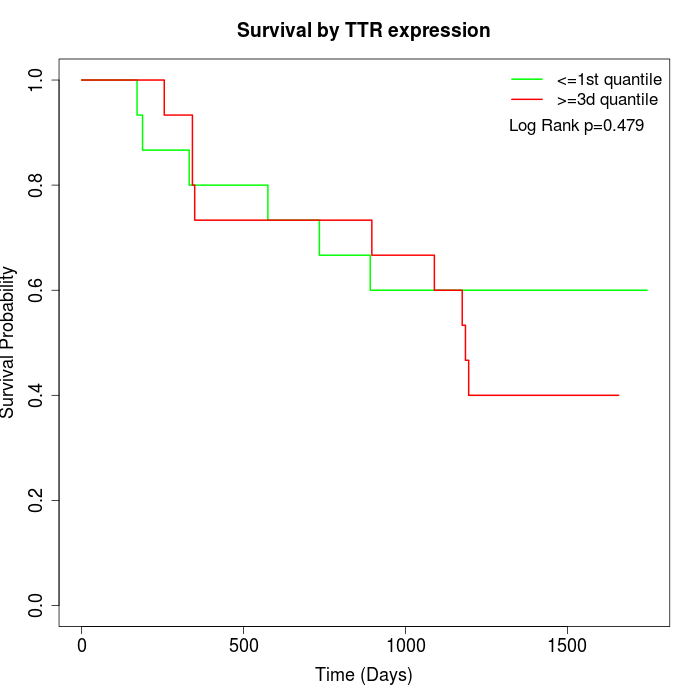
Survival by TTR expression:
Note: Click image to view full size file.
Copy number change of TTR:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TTR | 7276 | 1 | 6 | 23 | |
| GSE20123 | TTR | 7276 | 1 | 6 | 23 | |
| GSE43470 | TTR | 7276 | 0 | 2 | 41 | |
| GSE46452 | TTR | 7276 | 1 | 25 | 33 | |
| GSE47630 | TTR | 7276 | 5 | 20 | 15 | |
| GSE54993 | TTR | 7276 | 7 | 1 | 62 | |
| GSE54994 | TTR | 7276 | 2 | 16 | 35 | |
| GSE60625 | TTR | 7276 | 0 | 4 | 7 | |
| GSE74703 | TTR | 7276 | 0 | 2 | 34 | |
| GSE74704 | TTR | 7276 | 0 | 5 | 15 | |
| TCGA | TTR | 7276 | 14 | 36 | 46 |
Total number of gains: 31; Total number of losses: 123; Total Number of normals: 334.
Somatic mutations of TTR:
Generating mutation plots.
Highly correlated genes for TTR:
Showing top 20/945 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TTR | RUNDC3A | 0.75565 | 3 | 0 | 3 |
| TTR | NCR2 | 0.751871 | 3 | 0 | 3 |
| TTR | GPR4 | 0.749808 | 3 | 0 | 3 |
| TTR | SMIM6 | 0.747364 | 4 | 0 | 4 |
| TTR | LCT | 0.744681 | 4 | 0 | 4 |
| TTR | NMUR2 | 0.709702 | 3 | 0 | 3 |
| TTR | MTAP | 0.706407 | 3 | 0 | 3 |
| TTR | KLK4 | 0.703218 | 3 | 0 | 3 |
| TTR | IFT122 | 0.703173 | 4 | 0 | 4 |
| TTR | OR1F1 | 0.701277 | 4 | 0 | 4 |
| TTR | SCNN1G | 0.699832 | 4 | 0 | 4 |
| TTR | CAMP | 0.697083 | 3 | 0 | 3 |
| TTR | HGF | 0.691785 | 4 | 0 | 4 |
| TTR | ADRA1B | 0.691487 | 3 | 0 | 3 |
| TTR | RNF17 | 0.683679 | 3 | 0 | 3 |
| TTR | RFX6 | 0.683456 | 3 | 0 | 3 |
| TTR | LGI2 | 0.681594 | 3 | 0 | 3 |
| TTR | EPOR | 0.680838 | 5 | 0 | 5 |
| TTR | PRDM11 | 0.680663 | 4 | 0 | 4 |
| TTR | IFNA17 | 0.679776 | 5 | 0 | 5 |
For details and further investigation, click here