| Full name: serine protease 33 | Alias Symbol: EOS | ||
| Type: protein-coding gene | Cytoband: 16p13.3 | ||
| Entrez ID: 260429 | HGNC ID: HGNC:30405 | Ensembl Gene: ENSG00000103355 | OMIM ID: 613797 |
| Drug and gene relationship at DGIdb | |||
Expression of PRSS33:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PRSS33 | 260429 | 1552348_at | -0.3057 | 0.2874 | |
| GSE26886 | PRSS33 | 260429 | 1552348_at | -0.1990 | 0.2060 | |
| GSE45670 | PRSS33 | 260429 | 1552349_a_at | -0.0740 | 0.5319 | |
| GSE53622 | PRSS33 | 260429 | 128512 | 0.0972 | 0.6689 | |
| GSE53624 | PRSS33 | 260429 | 128512 | 0.0223 | 0.8820 | |
| GSE63941 | PRSS33 | 260429 | 1552348_at | 0.6032 | 0.1688 | |
| GSE77861 | PRSS33 | 260429 | 1552348_at | -0.1326 | 0.1256 | |
| TCGA | PRSS33 | 260429 | RNAseq | 1.2538 | 0.2837 |
Upregulated datasets: 0; Downregulated datasets: 0.
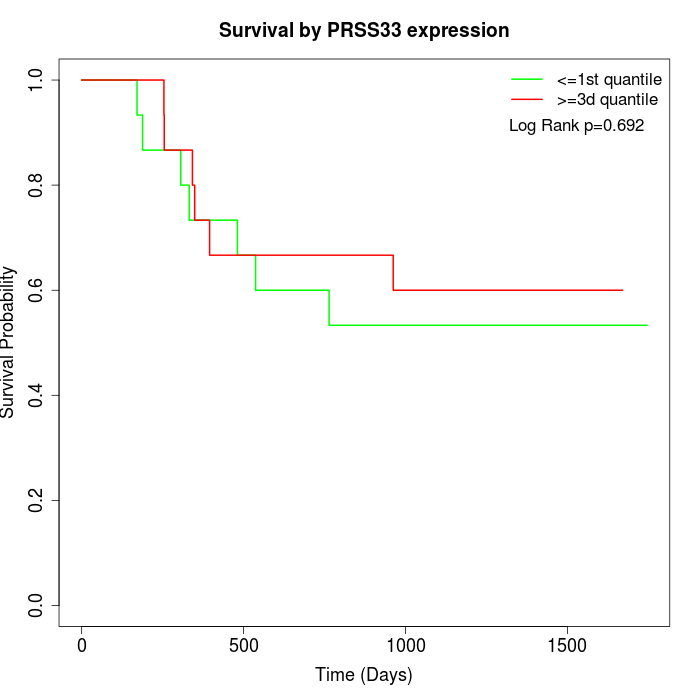
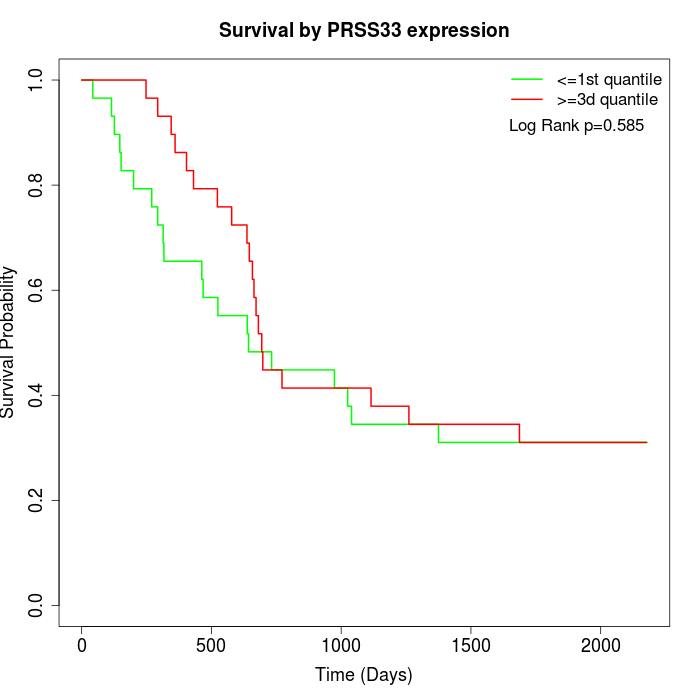
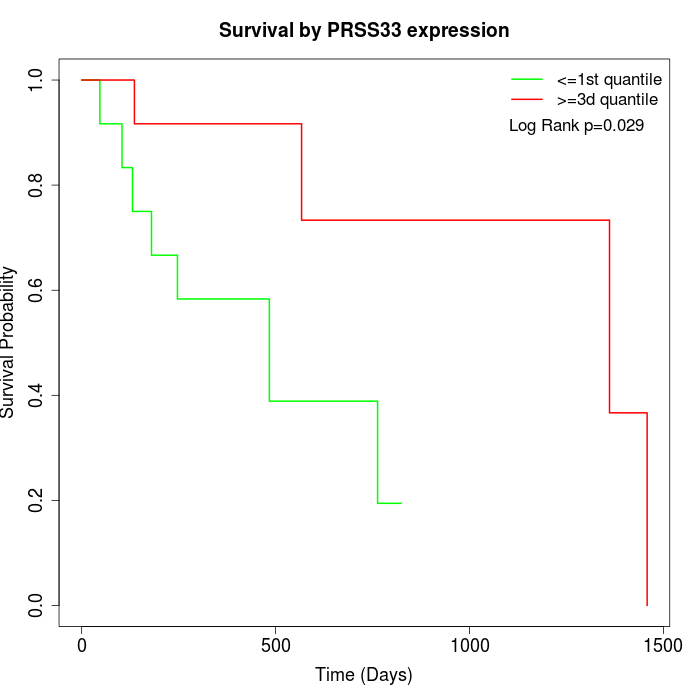
Survival by PRSS33 expression:
Note: Click image to view full size file.
Copy number change of PRSS33:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PRSS33 | 260429 | 5 | 5 | 20 | |
| GSE20123 | PRSS33 | 260429 | 5 | 4 | 21 | |
| GSE43470 | PRSS33 | 260429 | 4 | 7 | 32 | |
| GSE46452 | PRSS33 | 260429 | 38 | 1 | 20 | |
| GSE47630 | PRSS33 | 260429 | 13 | 6 | 21 | |
| GSE54993 | PRSS33 | 260429 | 3 | 5 | 62 | |
| GSE54994 | PRSS33 | 260429 | 5 | 9 | 39 | |
| GSE60625 | PRSS33 | 260429 | 4 | 0 | 7 | |
| GSE74703 | PRSS33 | 260429 | 4 | 5 | 27 | |
| GSE74704 | PRSS33 | 260429 | 3 | 2 | 15 | |
| TCGA | PRSS33 | 260429 | 19 | 13 | 64 |
Total number of gains: 103; Total number of losses: 57; Total Number of normals: 328.
Somatic mutations of PRSS33:
Generating mutation plots.
Highly correlated genes for PRSS33:
Showing top 20/198 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PRSS33 | CRIP2 | 0.755113 | 3 | 0 | 3 |
| PRSS33 | SMR3B | 0.724203 | 3 | 0 | 3 |
| PRSS33 | TAC3 | 0.708391 | 4 | 0 | 3 |
| PRSS33 | OMP | 0.700419 | 3 | 0 | 3 |
| PRSS33 | DSCR10 | 0.69977 | 3 | 0 | 3 |
| PRSS33 | HPSE2 | 0.693846 | 3 | 0 | 3 |
| PRSS33 | RAB4B | 0.683823 | 4 | 0 | 3 |
| PRSS33 | E2F4 | 0.678285 | 5 | 0 | 4 |
| PRSS33 | ABHD14B | 0.674288 | 3 | 0 | 3 |
| PRSS33 | TTR | 0.671789 | 3 | 0 | 3 |
| PRSS33 | HTR1E | 0.671347 | 3 | 0 | 3 |
| PRSS33 | KRTAP10-11 | 0.670474 | 3 | 0 | 3 |
| PRSS33 | SEMA5B | 0.669629 | 3 | 0 | 3 |
| PRSS33 | SIRT3 | 0.665444 | 4 | 0 | 4 |
| PRSS33 | SAMD15 | 0.661618 | 3 | 0 | 3 |
| PRSS33 | CEACAM21 | 0.657562 | 3 | 0 | 3 |
| PRSS33 | NLRP8 | 0.65719 | 5 | 0 | 4 |
| PRSS33 | PF4 | 0.656952 | 3 | 0 | 3 |
| PRSS33 | PKNOX2 | 0.656864 | 4 | 0 | 4 |
| PRSS33 | CCDC13 | 0.655928 | 5 | 0 | 4 |
For details and further investigation, click here