| Full name: glycosyltransferase 1 domain containing 1 | Alias Symbol: FLJ31978 | ||
| Type: protein-coding gene | Cytoband: 12q24.33 | ||
| Entrez ID: 144423 | HGNC ID: HGNC:26483 | Ensembl Gene: ENSG00000151948 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of GLT1D1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GLT1D1 | 144423 | 229770_at | -0.1351 | 0.7980 | |
| GSE26886 | GLT1D1 | 144423 | 229770_at | 0.0274 | 0.9112 | |
| GSE45670 | GLT1D1 | 144423 | 229770_at | 0.1620 | 0.2569 | |
| GSE53622 | GLT1D1 | 144423 | 101920 | 0.3820 | 0.0314 | |
| GSE53624 | GLT1D1 | 144423 | 101920 | 0.1020 | 0.5115 | |
| GSE63941 | GLT1D1 | 144423 | 229770_at | 0.4325 | 0.1180 | |
| GSE77861 | GLT1D1 | 144423 | 229770_at | -0.0404 | 0.7645 | |
| GSE97050 | GLT1D1 | 144423 | A_32_P19294 | -0.1017 | 0.8248 | |
| SRP219564 | GLT1D1 | 144423 | RNAseq | 1.3537 | 0.0446 | |
| TCGA | GLT1D1 | 144423 | RNAseq | 0.0372 | 0.9297 |
Upregulated datasets: 1; Downregulated datasets: 0.
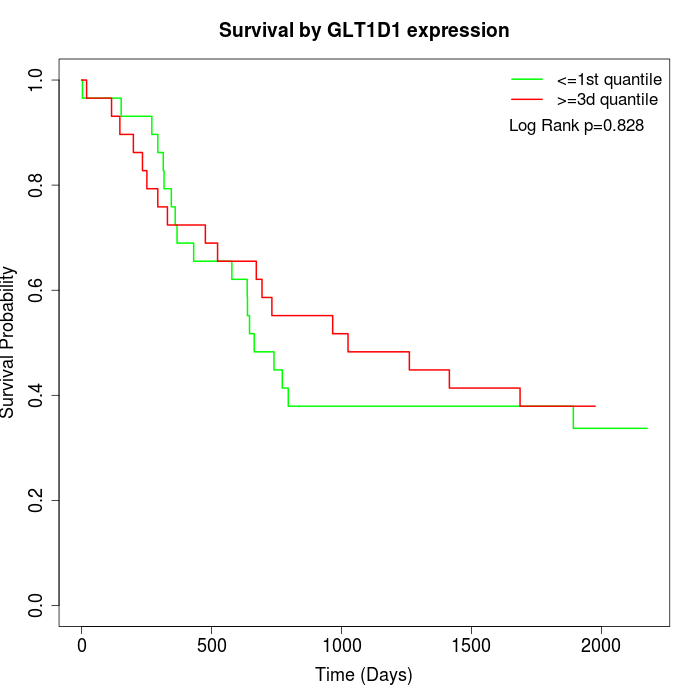
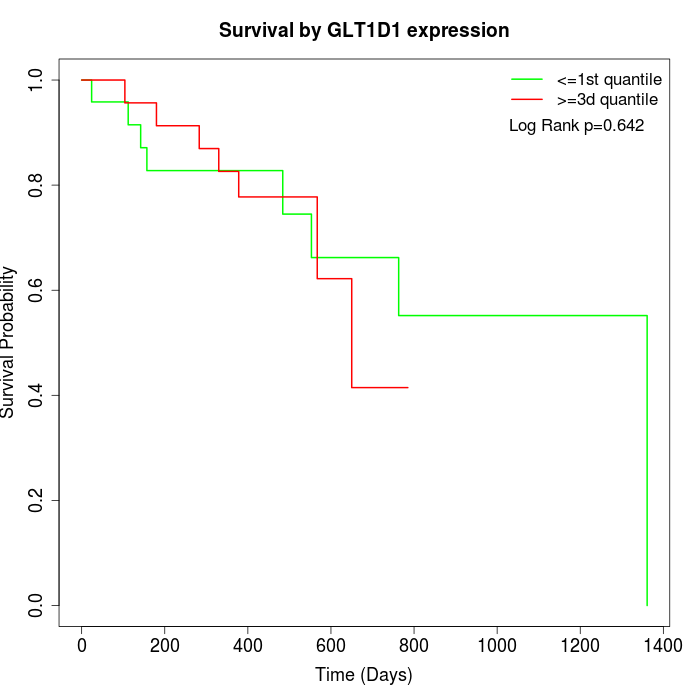
Survival by GLT1D1 expression:
Note: Click image to view full size file.
Copy number change of GLT1D1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GLT1D1 | 144423 | 3 | 4 | 23 | |
| GSE20123 | GLT1D1 | 144423 | 3 | 4 | 23 | |
| GSE43470 | GLT1D1 | 144423 | 2 | 0 | 41 | |
| GSE46452 | GLT1D1 | 144423 | 9 | 1 | 49 | |
| GSE47630 | GLT1D1 | 144423 | 9 | 3 | 28 | |
| GSE54993 | GLT1D1 | 144423 | 0 | 5 | 65 | |
| GSE54994 | GLT1D1 | 144423 | 4 | 7 | 42 | |
| GSE60625 | GLT1D1 | 144423 | 0 | 0 | 11 | |
| GSE74703 | GLT1D1 | 144423 | 2 | 0 | 34 | |
| GSE74704 | GLT1D1 | 144423 | 2 | 3 | 15 | |
| TCGA | GLT1D1 | 144423 | 22 | 14 | 60 |
Total number of gains: 56; Total number of losses: 41; Total Number of normals: 391.
Somatic mutations of GLT1D1:
Generating mutation plots.
Highly correlated genes for GLT1D1:
Showing top 20/37 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GLT1D1 | TNFRSF10C | 0.70052 | 3 | 0 | 3 |
| GLT1D1 | NMUR2 | 0.656666 | 3 | 0 | 3 |
| GLT1D1 | CMTM2 | 0.641542 | 4 | 0 | 3 |
| GLT1D1 | CEACAM4 | 0.630054 | 4 | 0 | 3 |
| GLT1D1 | AQP9 | 0.62672 | 4 | 0 | 4 |
| GLT1D1 | SPEM1 | 0.617761 | 3 | 0 | 3 |
| GLT1D1 | ACACB | 0.61214 | 3 | 0 | 3 |
| GLT1D1 | E2F4 | 0.607288 | 3 | 0 | 3 |
| GLT1D1 | PLEK | 0.605491 | 3 | 0 | 3 |
| GLT1D1 | TPSG1 | 0.59628 | 4 | 0 | 3 |
| GLT1D1 | CXCR1 | 0.590241 | 6 | 0 | 3 |
| GLT1D1 | GLRA2 | 0.580402 | 3 | 0 | 3 |
| GLT1D1 | KCNK17 | 0.578774 | 3 | 0 | 3 |
| GLT1D1 | SYT15 | 0.576388 | 3 | 0 | 3 |
| GLT1D1 | ATOH1 | 0.574887 | 4 | 0 | 3 |
| GLT1D1 | GAMT | 0.568546 | 4 | 0 | 3 |
| GLT1D1 | PROK2 | 0.568073 | 6 | 0 | 4 |
| GLT1D1 | RNF181 | 0.565546 | 3 | 0 | 3 |
| GLT1D1 | LINC01312 | 0.564961 | 3 | 0 | 3 |
| GLT1D1 | GLS2 | 0.557034 | 3 | 0 | 3 |
For details and further investigation, click here