| Full name: NADPH oxidase 1 | Alias Symbol: NOH1|NOH-1|MOX1|GP91-2 | ||
| Type: protein-coding gene | Cytoband: Xq22.1 | ||
| Entrez ID: 27035 | HGNC ID: HGNC:7889 | Ensembl Gene: ENSG00000007952 | OMIM ID: 300225 |
| Drug and gene relationship at DGIdb | |||
NOX1 involved pathways:
Expression of NOX1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NOX1 | 27035 | 210808_s_at | 0.0091 | 0.9726 | |
| GSE20347 | NOX1 | 27035 | 207217_s_at | 0.0757 | 0.3532 | |
| GSE23400 | NOX1 | 27035 | 207380_x_at | -0.0109 | 0.7987 | |
| GSE26886 | NOX1 | 27035 | 210808_s_at | 0.2609 | 0.0078 | |
| GSE29001 | NOX1 | 27035 | 207217_s_at | -0.1282 | 0.4686 | |
| GSE38129 | NOX1 | 27035 | 210808_s_at | -0.1152 | 0.0445 | |
| GSE45670 | NOX1 | 27035 | 207380_x_at | 0.1697 | 0.0592 | |
| GSE53622 | NOX1 | 27035 | 1275 | -0.2543 | 0.0960 | |
| GSE53624 | NOX1 | 27035 | 1275 | -0.4683 | 0.0001 | |
| GSE63941 | NOX1 | 27035 | 210808_s_at | -0.0233 | 0.8810 | |
| GSE77861 | NOX1 | 27035 | 210808_s_at | -0.0720 | 0.5271 | |
| GSE97050 | NOX1 | 27035 | A_33_P3286552 | 0.0283 | 0.8996 | |
| SRP219564 | NOX1 | 27035 | RNAseq | 0.6777 | 0.1092 | |
| TCGA | NOX1 | 27035 | RNAseq | 1.2605 | 0.0114 |
Upregulated datasets: 1; Downregulated datasets: 0.
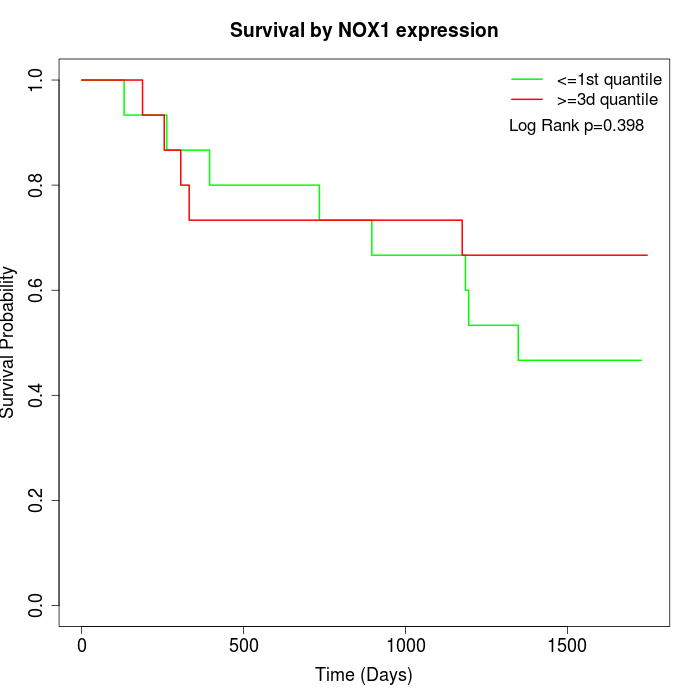
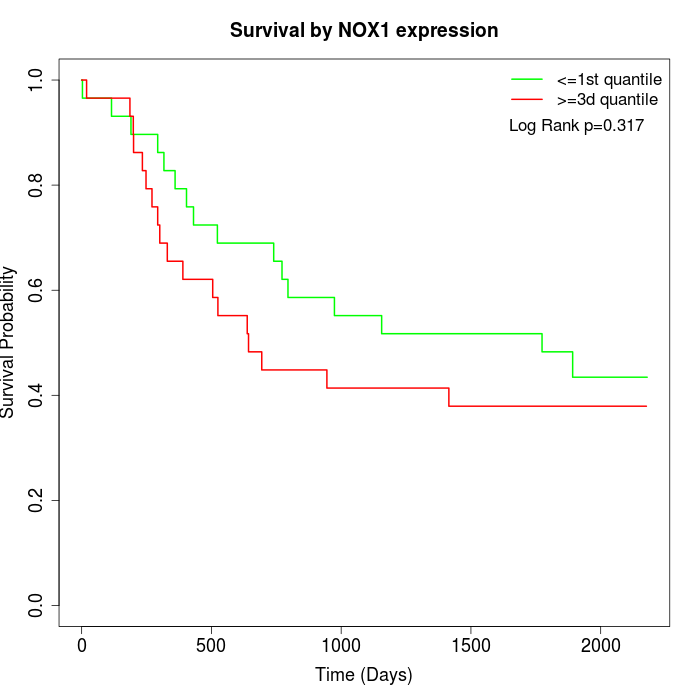
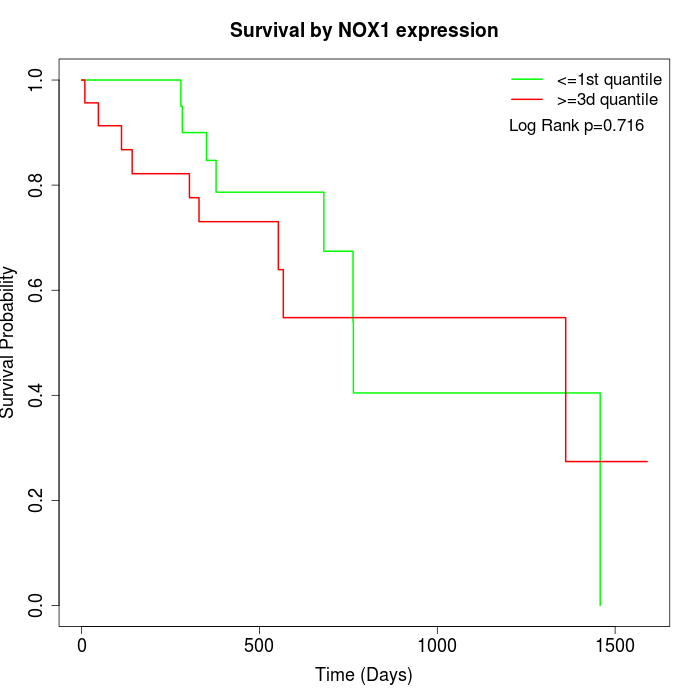
Survival by NOX1 expression:
Note: Click image to view full size file.
Copy number change of NOX1:
No record found for this gene.
Somatic mutations of NOX1:
Generating mutation plots.
Highly correlated genes for NOX1:
Showing top 20/531 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NOX1 | OSBPL5 | 0.825765 | 3 | 0 | 3 |
| NOX1 | MGAT5B | 0.798326 | 3 | 0 | 3 |
| NOX1 | ATAD3C | 0.795707 | 3 | 0 | 3 |
| NOX1 | LIMD1 | 0.794716 | 3 | 0 | 3 |
| NOX1 | CDH10 | 0.78306 | 4 | 0 | 4 |
| NOX1 | TRAPPC9 | 0.782372 | 4 | 0 | 4 |
| NOX1 | SLC5A4 | 0.777826 | 4 | 0 | 4 |
| NOX1 | PHGR1 | 0.758515 | 3 | 0 | 3 |
| NOX1 | KIR2DS5 | 0.735651 | 4 | 0 | 4 |
| NOX1 | IFNA4 | 0.734241 | 5 | 0 | 5 |
| NOX1 | NRIP2 | 0.733973 | 4 | 0 | 4 |
| NOX1 | ATXN8OS | 0.733109 | 4 | 0 | 4 |
| NOX1 | NTNG1 | 0.730593 | 3 | 0 | 3 |
| NOX1 | NCR1 | 0.728481 | 4 | 0 | 4 |
| NOX1 | KLK2 | 0.725891 | 5 | 0 | 4 |
| NOX1 | TTC9B | 0.724385 | 3 | 0 | 3 |
| NOX1 | DUSP9 | 0.720868 | 3 | 0 | 3 |
| NOX1 | STK32C | 0.714433 | 3 | 0 | 3 |
| NOX1 | PRX | 0.714032 | 5 | 0 | 5 |
| NOX1 | DUSP15 | 0.713243 | 3 | 0 | 3 |
For details and further investigation, click here