| Full name: LIM domains containing 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 3p21.31 | ||
| Entrez ID: 8994 | HGNC ID: HGNC:6612 | Ensembl Gene: ENSG00000144791 | OMIM ID: 604543 |
| Drug and gene relationship at DGIdb | |||
LIMD1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04390 | Hippo signaling pathway |
Expression of LIMD1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LIMD1 | 8994 | 222762_x_at | 0.3906 | 0.3824 | |
| GSE20347 | LIMD1 | 8994 | 218850_s_at | -0.0112 | 0.8929 | |
| GSE23400 | LIMD1 | 8994 | 218850_s_at | -0.1227 | 0.0115 | |
| GSE26886 | LIMD1 | 8994 | 234687_x_at | -0.0038 | 0.9755 | |
| GSE29001 | LIMD1 | 8994 | 218850_s_at | -0.0841 | 0.6372 | |
| GSE38129 | LIMD1 | 8994 | 218850_s_at | -0.0615 | 0.5696 | |
| GSE45670 | LIMD1 | 8994 | 222762_x_at | -0.0136 | 0.9363 | |
| GSE53622 | LIMD1 | 8994 | 11543 | -0.3140 | 0.0001 | |
| GSE53624 | LIMD1 | 8994 | 98482 | -0.1113 | 0.1538 | |
| GSE63941 | LIMD1 | 8994 | 222762_x_at | -0.7672 | 0.0454 | |
| GSE77861 | LIMD1 | 8994 | 234687_x_at | -0.1451 | 0.1761 | |
| GSE97050 | LIMD1 | 8994 | A_33_P3276053 | -0.1262 | 0.5922 | |
| SRP007169 | LIMD1 | 8994 | RNAseq | -0.0029 | 0.9947 | |
| SRP008496 | LIMD1 | 8994 | RNAseq | -0.1319 | 0.6194 | |
| SRP064894 | LIMD1 | 8994 | RNAseq | -0.0998 | 0.4422 | |
| SRP133303 | LIMD1 | 8994 | RNAseq | -0.8469 | 0.0000 | |
| SRP159526 | LIMD1 | 8994 | RNAseq | 0.0417 | 0.8124 | |
| SRP193095 | LIMD1 | 8994 | RNAseq | -0.0992 | 0.3347 | |
| SRP219564 | LIMD1 | 8994 | RNAseq | -0.5087 | 0.1315 | |
| TCGA | LIMD1 | 8994 | RNAseq | -0.2145 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
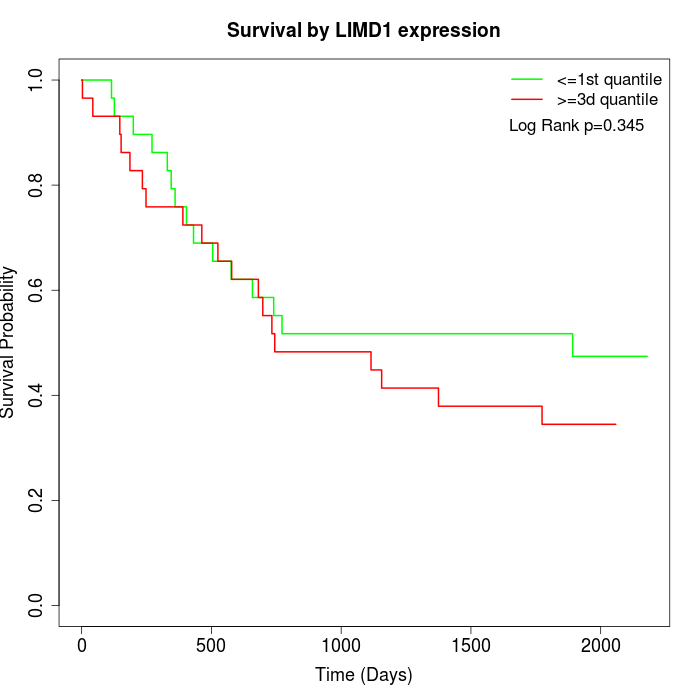
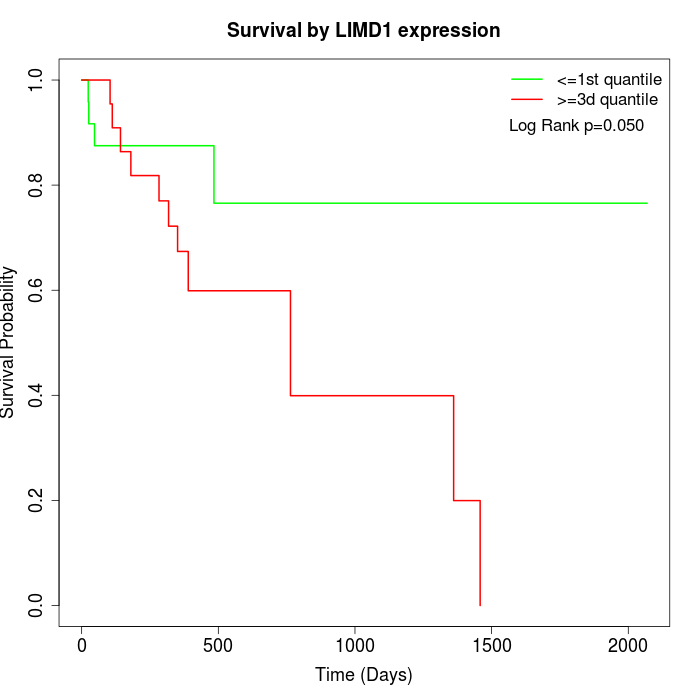
Survival by LIMD1 expression:
Note: Click image to view full size file.
Copy number change of LIMD1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LIMD1 | 8994 | 0 | 17 | 13 | |
| GSE20123 | LIMD1 | 8994 | 0 | 18 | 12 | |
| GSE43470 | LIMD1 | 8994 | 0 | 20 | 23 | |
| GSE46452 | LIMD1 | 8994 | 2 | 17 | 40 | |
| GSE47630 | LIMD1 | 8994 | 1 | 24 | 15 | |
| GSE54993 | LIMD1 | 8994 | 7 | 2 | 61 | |
| GSE54994 | LIMD1 | 8994 | 0 | 36 | 17 | |
| GSE60625 | LIMD1 | 8994 | 5 | 0 | 6 | |
| GSE74703 | LIMD1 | 8994 | 0 | 15 | 21 | |
| GSE74704 | LIMD1 | 8994 | 0 | 12 | 8 | |
| TCGA | LIMD1 | 8994 | 2 | 74 | 20 |
Total number of gains: 17; Total number of losses: 235; Total Number of normals: 236.
Somatic mutations of LIMD1:
Generating mutation plots.
Highly correlated genes for LIMD1:
Showing top 20/121 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LIMD1 | GPSM3 | 0.823352 | 3 | 0 | 3 |
| LIMD1 | TRPM3 | 0.81971 | 3 | 0 | 3 |
| LIMD1 | LMAN1L | 0.809395 | 3 | 0 | 3 |
| LIMD1 | NOX1 | 0.794716 | 3 | 0 | 3 |
| LIMD1 | KLHL35 | 0.787487 | 4 | 0 | 3 |
| LIMD1 | KIR3DL3 | 0.785156 | 3 | 0 | 3 |
| LIMD1 | ZNF283 | 0.775002 | 3 | 0 | 3 |
| LIMD1 | B3GALT1 | 0.770434 | 3 | 0 | 3 |
| LIMD1 | RNF17 | 0.758679 | 3 | 0 | 3 |
| LIMD1 | SLC19A3 | 0.748282 | 3 | 0 | 3 |
| LIMD1 | LRCH4 | 0.729099 | 3 | 0 | 3 |
| LIMD1 | NICN1 | 0.722975 | 3 | 0 | 3 |
| LIMD1 | PTPRN2 | 0.713172 | 5 | 0 | 4 |
| LIMD1 | TLR6 | 0.711961 | 3 | 0 | 3 |
| LIMD1 | ADORA3 | 0.71108 | 3 | 0 | 3 |
| LIMD1 | FFAR2 | 0.709339 | 3 | 0 | 3 |
| LIMD1 | SSH3 | 0.708431 | 4 | 0 | 4 |
| LIMD1 | SLC24A4 | 0.700132 | 3 | 0 | 3 |
| LIMD1 | FSHR | 0.698024 | 3 | 0 | 3 |
| LIMD1 | TP53INP1 | 0.697801 | 3 | 0 | 3 |
For details and further investigation, click here