| Full name: phosphoglycerate mutase 1 | Alias Symbol: PGAM-B | ||
| Type: protein-coding gene | Cytoband: 10q24.1 | ||
| Entrez ID: 5223 | HGNC ID: HGNC:8888 | Ensembl Gene: ENSG00000171314 | OMIM ID: 172250 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of PGAM1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PGAM1 | 5223 | 200886_s_at | -0.2138 | 0.5181 | |
| GSE20347 | PGAM1 | 5223 | 200886_s_at | -0.5132 | 0.0005 | |
| GSE23400 | PGAM1 | 5223 | 200886_s_at | -0.2332 | 0.0001 | |
| GSE26886 | PGAM1 | 5223 | 200886_s_at | -0.6674 | 0.0001 | |
| GSE29001 | PGAM1 | 5223 | 200886_s_at | -0.3600 | 0.0751 | |
| GSE38129 | PGAM1 | 5223 | 200886_s_at | -0.2141 | 0.1386 | |
| GSE45670 | PGAM1 | 5223 | 200886_s_at | -0.0069 | 0.9653 | |
| GSE53622 | PGAM1 | 5223 | 46526 | -0.3706 | 0.0001 | |
| GSE53624 | PGAM1 | 5223 | 46526 | -0.3893 | 0.0000 | |
| GSE63941 | PGAM1 | 5223 | 200886_s_at | -0.1501 | 0.7020 | |
| GSE77861 | PGAM1 | 5223 | 200886_s_at | -0.2535 | 0.3421 | |
| GSE97050 | PGAM1 | 5223 | A_33_P3268334 | -0.2838 | 0.3418 | |
| SRP007169 | PGAM1 | 5223 | RNAseq | -1.4247 | 0.0000 | |
| SRP008496 | PGAM1 | 5223 | RNAseq | -1.3131 | 0.0000 | |
| SRP064894 | PGAM1 | 5223 | RNAseq | -0.4597 | 0.0032 | |
| SRP133303 | PGAM1 | 5223 | RNAseq | -0.0513 | 0.7729 | |
| SRP159526 | PGAM1 | 5223 | RNAseq | -0.6978 | 0.1255 | |
| SRP193095 | PGAM1 | 5223 | RNAseq | -0.3849 | 0.0579 | |
| SRP219564 | PGAM1 | 5223 | RNAseq | -0.8628 | 0.0076 | |
| TCGA | PGAM1 | 5223 | RNAseq | 0.1571 | 0.0033 |
Upregulated datasets: 0; Downregulated datasets: 2.
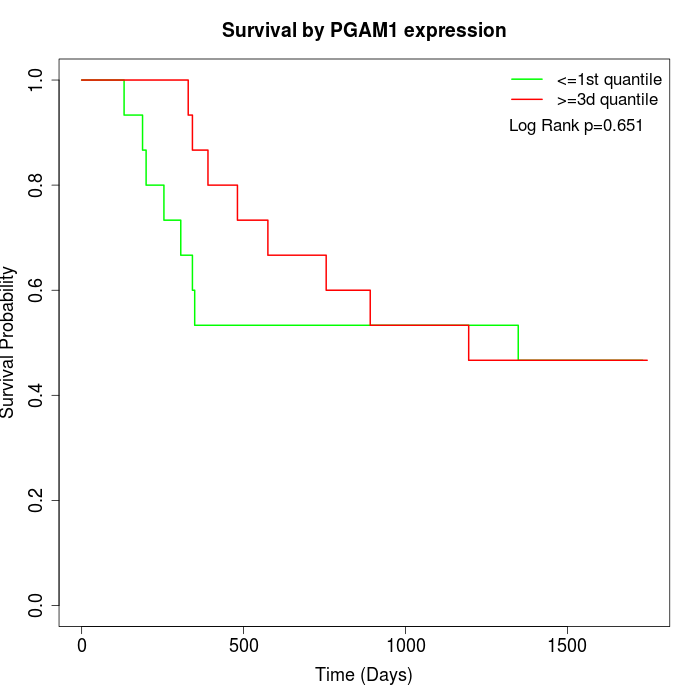
Survival by PGAM1 expression:
Note: Click image to view full size file.
Copy number change of PGAM1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PGAM1 | 5223 | 0 | 8 | 22 | |
| GSE20123 | PGAM1 | 5223 | 0 | 7 | 23 | |
| GSE43470 | PGAM1 | 5223 | 0 | 7 | 36 | |
| GSE46452 | PGAM1 | 5223 | 0 | 11 | 48 | |
| GSE47630 | PGAM1 | 5223 | 2 | 14 | 24 | |
| GSE54993 | PGAM1 | 5223 | 8 | 0 | 62 | |
| GSE54994 | PGAM1 | 5223 | 2 | 10 | 41 | |
| GSE60625 | PGAM1 | 5223 | 0 | 0 | 11 | |
| GSE74703 | PGAM1 | 5223 | 0 | 5 | 31 | |
| GSE74704 | PGAM1 | 5223 | 0 | 4 | 16 | |
| TCGA | PGAM1 | 5223 | 5 | 28 | 63 |
Total number of gains: 17; Total number of losses: 94; Total Number of normals: 377.
Somatic mutations of PGAM1:
Generating mutation plots.
Highly correlated genes for PGAM1:
Showing top 20/887 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PGAM1 | SPNS2 | 0.720763 | 3 | 0 | 3 |
| PGAM1 | BTBD11 | 0.716694 | 4 | 0 | 4 |
| PGAM1 | ORMDL2 | 0.712927 | 9 | 0 | 9 |
| PGAM1 | TBC1D20 | 0.707145 | 3 | 0 | 3 |
| PGAM1 | ZNF709 | 0.701445 | 3 | 0 | 3 |
| PGAM1 | CCDC25 | 0.691073 | 3 | 0 | 3 |
| PGAM1 | CCDC85C | 0.690762 | 4 | 0 | 4 |
| PGAM1 | LYSMD3 | 0.682009 | 3 | 0 | 3 |
| PGAM1 | MAB21L3 | 0.679263 | 4 | 0 | 4 |
| PGAM1 | XKRX | 0.674226 | 4 | 0 | 3 |
| PGAM1 | VDAC2 | 0.673791 | 11 | 0 | 10 |
| PGAM1 | FAR1 | 0.672214 | 3 | 0 | 3 |
| PGAM1 | VPS25 | 0.67013 | 6 | 0 | 6 |
| PGAM1 | PDCD6IP | 0.669984 | 9 | 0 | 8 |
| PGAM1 | SNX14 | 0.667622 | 3 | 0 | 3 |
| PGAM1 | PRSS27 | 0.660632 | 4 | 0 | 4 |
| PGAM1 | MIR210HG | 0.660062 | 7 | 0 | 6 |
| PGAM1 | LRRC28 | 0.6596 | 3 | 0 | 3 |
| PGAM1 | A2ML1 | 0.65807 | 4 | 0 | 4 |
| PGAM1 | SH3GLB1 | 0.657028 | 11 | 0 | 10 |
For details and further investigation, click here