| Full name: SH3 domain containing GRB2 like, endophilin B1 | Alias Symbol: CGI-61|KIAA0491|Bif-1|PPP1R70 | ||
| Type: protein-coding gene | Cytoband: 1p22.3 | ||
| Entrez ID: 51100 | HGNC ID: HGNC:10833 | Ensembl Gene: ENSG00000097033 | OMIM ID: 609287 |
| Drug and gene relationship at DGIdb | |||
Expression of SH3GLB1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SH3GLB1 | 51100 | 209091_s_at | -0.8864 | 0.1134 | |
| GSE20347 | SH3GLB1 | 51100 | 209091_s_at | -1.6922 | 0.0000 | |
| GSE23400 | SH3GLB1 | 51100 | 210101_x_at | -0.8753 | 0.0000 | |
| GSE26886 | SH3GLB1 | 51100 | 209091_s_at | -1.9626 | 0.0000 | |
| GSE29001 | SH3GLB1 | 51100 | 209090_s_at | -0.5443 | 0.1005 | |
| GSE38129 | SH3GLB1 | 51100 | 209091_s_at | -0.9931 | 0.0000 | |
| GSE45670 | SH3GLB1 | 51100 | 209091_s_at | -0.4171 | 0.0674 | |
| GSE53622 | SH3GLB1 | 51100 | 19542 | -0.9344 | 0.0000 | |
| GSE53624 | SH3GLB1 | 51100 | 19542 | -0.9741 | 0.0000 | |
| GSE63941 | SH3GLB1 | 51100 | 209091_s_at | -0.5378 | 0.2159 | |
| GSE77861 | SH3GLB1 | 51100 | 210101_x_at | -1.0712 | 0.0010 | |
| GSE97050 | SH3GLB1 | 51100 | A_33_P3266315 | -0.7419 | 0.0835 | |
| SRP007169 | SH3GLB1 | 51100 | RNAseq | -1.3970 | 0.0002 | |
| SRP008496 | SH3GLB1 | 51100 | RNAseq | -1.1280 | 0.0000 | |
| SRP064894 | SH3GLB1 | 51100 | RNAseq | -0.7470 | 0.0000 | |
| SRP133303 | SH3GLB1 | 51100 | RNAseq | -0.2850 | 0.0695 | |
| SRP159526 | SH3GLB1 | 51100 | RNAseq | -1.5810 | 0.0000 | |
| SRP193095 | SH3GLB1 | 51100 | RNAseq | -0.8939 | 0.0000 | |
| SRP219564 | SH3GLB1 | 51100 | RNAseq | -0.6738 | 0.0401 | |
| TCGA | SH3GLB1 | 51100 | RNAseq | -0.0735 | 0.0824 |
Upregulated datasets: 0; Downregulated datasets: 6.
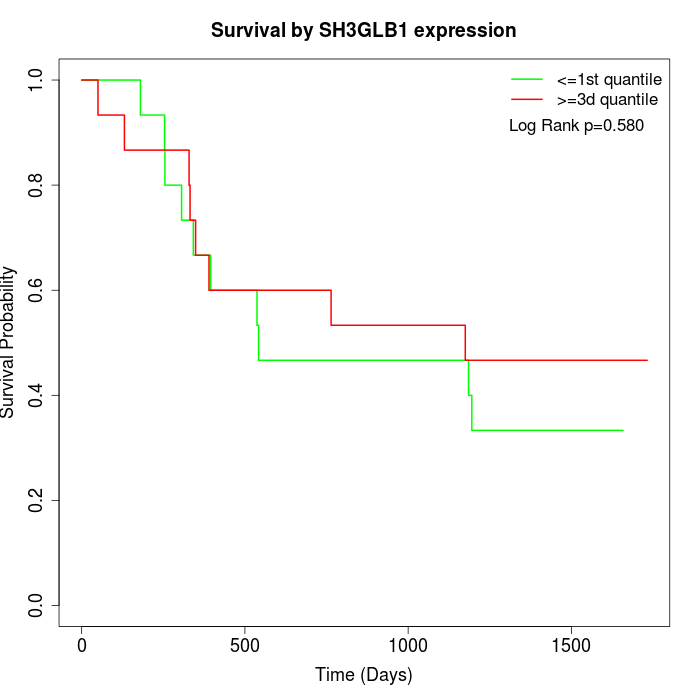
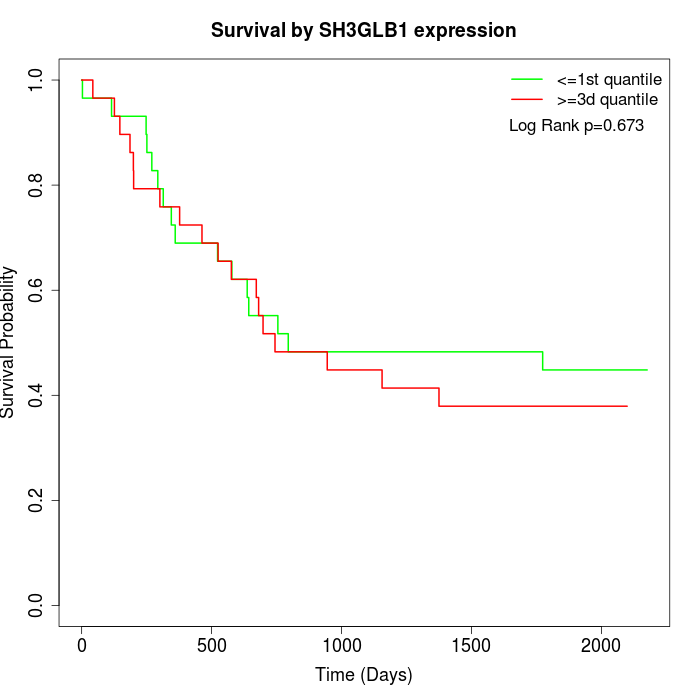
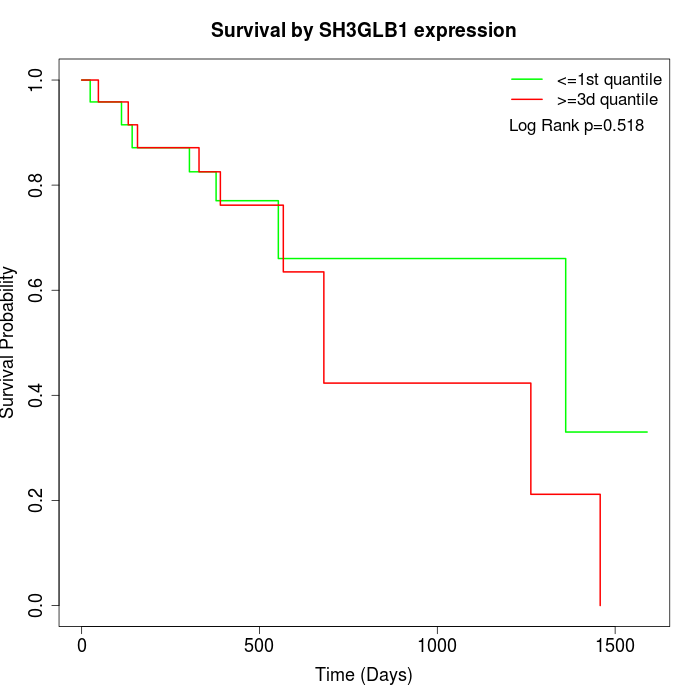
Survival by SH3GLB1 expression:
Note: Click image to view full size file.
Copy number change of SH3GLB1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SH3GLB1 | 51100 | 0 | 8 | 22 | |
| GSE20123 | SH3GLB1 | 51100 | 0 | 8 | 22 | |
| GSE43470 | SH3GLB1 | 51100 | 2 | 2 | 39 | |
| GSE46452 | SH3GLB1 | 51100 | 1 | 1 | 57 | |
| GSE47630 | SH3GLB1 | 51100 | 8 | 5 | 27 | |
| GSE54993 | SH3GLB1 | 51100 | 0 | 1 | 69 | |
| GSE54994 | SH3GLB1 | 51100 | 6 | 3 | 44 | |
| GSE60625 | SH3GLB1 | 51100 | 0 | 0 | 11 | |
| GSE74703 | SH3GLB1 | 51100 | 1 | 2 | 33 | |
| GSE74704 | SH3GLB1 | 51100 | 0 | 5 | 15 | |
| TCGA | SH3GLB1 | 51100 | 6 | 23 | 67 |
Total number of gains: 24; Total number of losses: 58; Total Number of normals: 406.
Somatic mutations of SH3GLB1:
Generating mutation plots.
Highly correlated genes for SH3GLB1:
Showing top 20/1779 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SH3GLB1 | FCHO2 | 0.857415 | 7 | 0 | 7 |
| SH3GLB1 | CCDC7 | 0.822482 | 3 | 0 | 3 |
| SH3GLB1 | SFTA2 | 0.8144 | 4 | 0 | 4 |
| SH3GLB1 | PLEKHA7 | 0.806754 | 7 | 0 | 7 |
| SH3GLB1 | FAM3D | 0.804277 | 6 | 0 | 6 |
| SH3GLB1 | TOM1 | 0.800417 | 10 | 0 | 10 |
| SH3GLB1 | ECM1 | 0.798386 | 10 | 0 | 10 |
| SH3GLB1 | RAB11A | 0.798159 | 11 | 0 | 10 |
| SH3GLB1 | ITCH | 0.796734 | 10 | 0 | 10 |
| SH3GLB1 | CYSRT1 | 0.793718 | 6 | 0 | 6 |
| SH3GLB1 | MAPK3 | 0.793006 | 9 | 0 | 9 |
| SH3GLB1 | SLC46A2 | 0.791563 | 7 | 0 | 7 |
| SH3GLB1 | NAGK | 0.788329 | 11 | 0 | 10 |
| SH3GLB1 | CCDC6 | 0.787028 | 10 | 0 | 10 |
| SH3GLB1 | CXCR2 | 0.785659 | 11 | 0 | 11 |
| SH3GLB1 | BRK1 | 0.783759 | 7 | 0 | 6 |
| SH3GLB1 | CAPN5 | 0.782153 | 11 | 0 | 10 |
| SH3GLB1 | ZFAND2B | 0.781278 | 7 | 0 | 7 |
| SH3GLB1 | RNF141 | 0.780315 | 11 | 0 | 10 |
| SH3GLB1 | PDCD6IP | 0.776336 | 10 | 0 | 9 |
For details and further investigation, click here