| Full name: serine protease 55 | Alias Symbol: T-SP1|UNQ9391|CT153 | ||
| Type: protein-coding gene | Cytoband: 8p23.1 | ||
| Entrez ID: 203074 | HGNC ID: HGNC:30824 | Ensembl Gene: ENSG00000184647 | OMIM ID: 615144 |
| Drug and gene relationship at DGIdb | |||
Expression of PRSS55:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PRSS55 | 203074 | 1570274_at | -0.1473 | 0.4389 | |
| GSE26886 | PRSS55 | 203074 | 1570274_at | -0.0840 | 0.4258 | |
| GSE45670 | PRSS55 | 203074 | 1570274_at | 0.0070 | 0.9366 | |
| GSE53622 | PRSS55 | 203074 | 78544 | 0.0333 | 0.7487 | |
| GSE53624 | PRSS55 | 203074 | 78544 | 0.0154 | 0.9100 | |
| GSE63941 | PRSS55 | 203074 | 1570274_at | 0.1327 | 0.3518 | |
| GSE77861 | PRSS55 | 203074 | 1570274_at | -0.0971 | 0.3591 | |
| TCGA | PRSS55 | 203074 | RNAseq | 1.6367 | 0.2654 |
Upregulated datasets: 0; Downregulated datasets: 0.
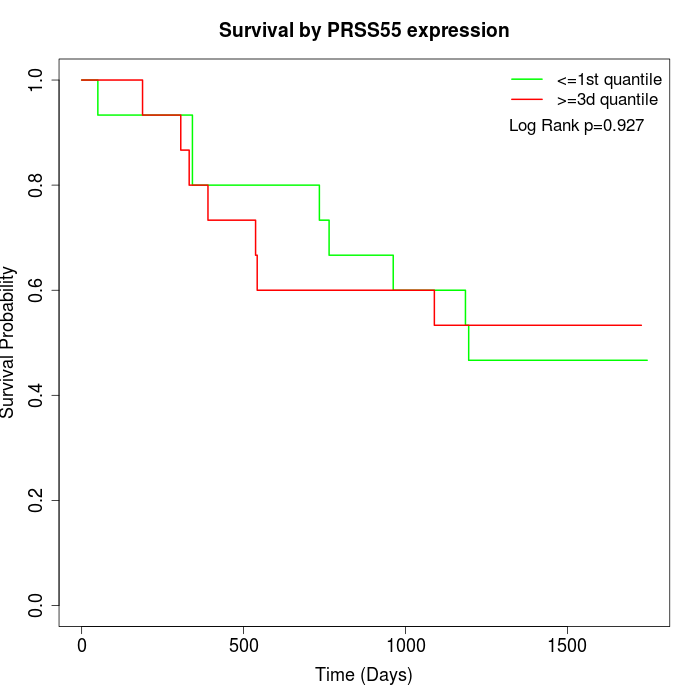
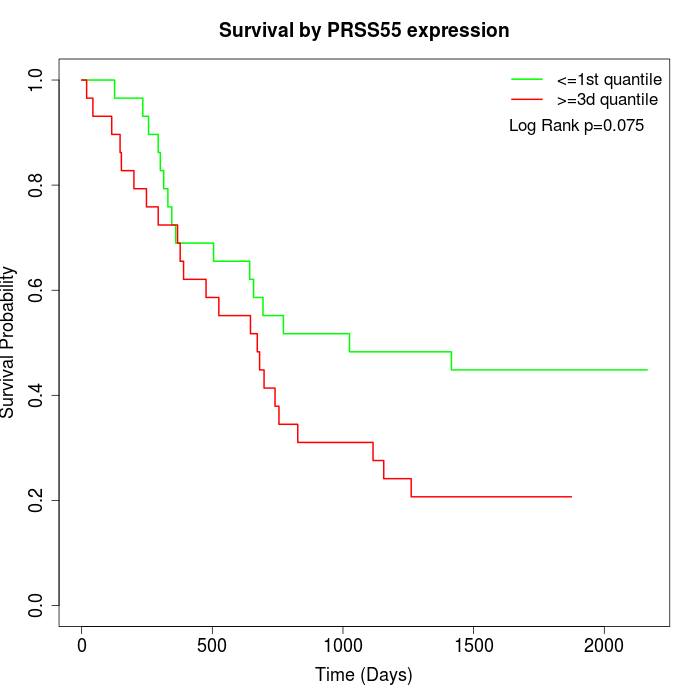
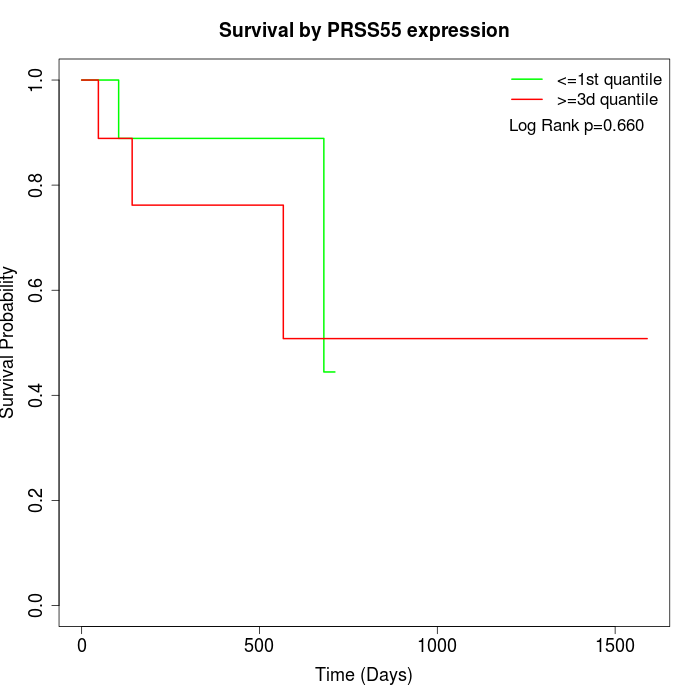
Survival by PRSS55 expression:
Note: Click image to view full size file.
Copy number change of PRSS55:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PRSS55 | 203074 | 4 | 11 | 15 | |
| GSE20123 | PRSS55 | 203074 | 4 | 11 | 15 | |
| GSE43470 | PRSS55 | 203074 | 4 | 9 | 30 | |
| GSE46452 | PRSS55 | 203074 | 12 | 14 | 33 | |
| GSE47630 | PRSS55 | 203074 | 10 | 7 | 23 | |
| GSE54993 | PRSS55 | 203074 | 3 | 14 | 53 | |
| GSE54994 | PRSS55 | 203074 | 9 | 19 | 25 | |
| GSE60625 | PRSS55 | 203074 | 3 | 0 | 8 | |
| GSE74703 | PRSS55 | 203074 | 4 | 7 | 25 | |
| GSE74704 | PRSS55 | 203074 | 3 | 7 | 10 | |
| TCGA | PRSS55 | 203074 | 13 | 40 | 43 |
Total number of gains: 69; Total number of losses: 139; Total Number of normals: 280.
Somatic mutations of PRSS55:
Generating mutation plots.
Highly correlated genes for PRSS55:
Showing top 20/42 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PRSS55 | RNASE7 | 0.665568 | 3 | 0 | 3 |
| PRSS55 | PHLDB3 | 0.664946 | 3 | 0 | 3 |
| PRSS55 | CHAD | 0.633255 | 3 | 0 | 3 |
| PRSS55 | LINC00424 | 0.63135 | 4 | 0 | 3 |
| PRSS55 | TMEM184A | 0.614861 | 4 | 0 | 3 |
| PRSS55 | LRRC29 | 0.61039 | 4 | 0 | 3 |
| PRSS55 | G6PC2 | 0.602911 | 3 | 0 | 3 |
| PRSS55 | SMPD2 | 0.599465 | 4 | 0 | 3 |
| PRSS55 | LINC01214 | 0.599263 | 4 | 0 | 3 |
| PRSS55 | MIR4755 | 0.590733 | 3 | 0 | 3 |
| PRSS55 | SLC22A14 | 0.57721 | 3 | 0 | 3 |
| PRSS55 | C2orf16 | 0.574721 | 3 | 0 | 3 |
| PRSS55 | KLHDC8A | 0.572434 | 3 | 0 | 3 |
| PRSS55 | NSDHL | 0.569569 | 3 | 0 | 3 |
| PRSS55 | S100G | 0.564853 | 3 | 0 | 3 |
| PRSS55 | GAL3ST3 | 0.564447 | 4 | 0 | 4 |
| PRSS55 | FANCD2OS | 0.562339 | 3 | 0 | 3 |
| PRSS55 | LINC00396 | 0.561218 | 4 | 0 | 3 |
| PRSS55 | NCR2 | 0.559021 | 4 | 0 | 4 |
| PRSS55 | LINC00589 | 0.550869 | 4 | 0 | 3 |
For details and further investigation, click here