| Full name: semenogelin 1 | Alias Symbol: CT103 | ||
| Type: protein-coding gene | Cytoband: 20q13.12 | ||
| Entrez ID: 6406 | HGNC ID: HGNC:10742 | Ensembl Gene: ENSG00000124233 | OMIM ID: 182140 |
| Drug and gene relationship at DGIdb | |||
Expression of SEMG1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SEMG1 | 6406 | 206442_at | -0.0591 | 0.8200 | |
| GSE20347 | SEMG1 | 6406 | 206442_at | -0.0684 | 0.2803 | |
| GSE23400 | SEMG1 | 6406 | 206442_at | -0.1063 | 0.0000 | |
| GSE26886 | SEMG1 | 6406 | 206442_at | -0.0725 | 0.2752 | |
| GSE29001 | SEMG1 | 6406 | 206442_at | -0.1201 | 0.3192 | |
| GSE38129 | SEMG1 | 6406 | 206442_at | -0.0362 | 0.4978 | |
| GSE45670 | SEMG1 | 6406 | 206442_at | 0.0829 | 0.3499 | |
| GSE53622 | SEMG1 | 6406 | 2241 | 0.3932 | 0.0013 | |
| GSE53624 | SEMG1 | 6406 | 2241 | 0.2971 | 0.0132 | |
| GSE63941 | SEMG1 | 6406 | 206442_at | 0.0453 | 0.8181 | |
| GSE77861 | SEMG1 | 6406 | 206442_at | -0.1125 | 0.1566 | |
| TCGA | SEMG1 | 6406 | RNAseq | -1.9358 | 0.2754 |
Upregulated datasets: 0; Downregulated datasets: 0.
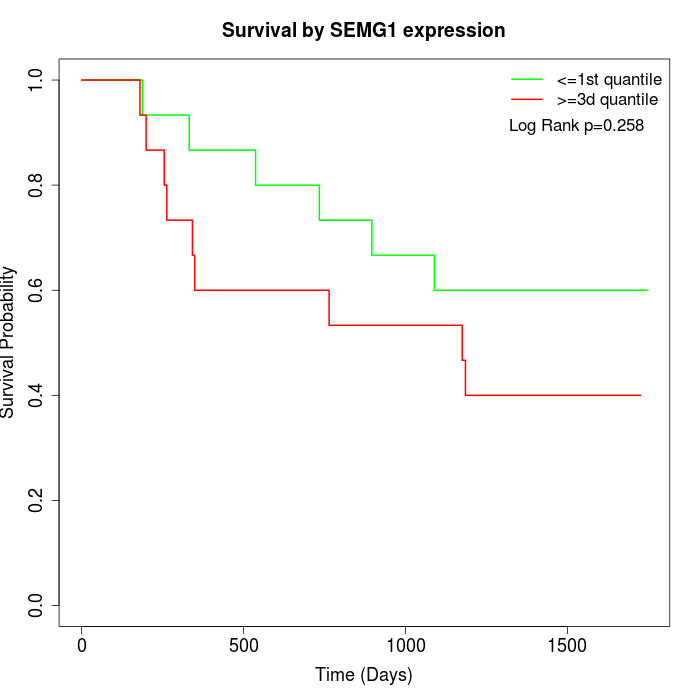
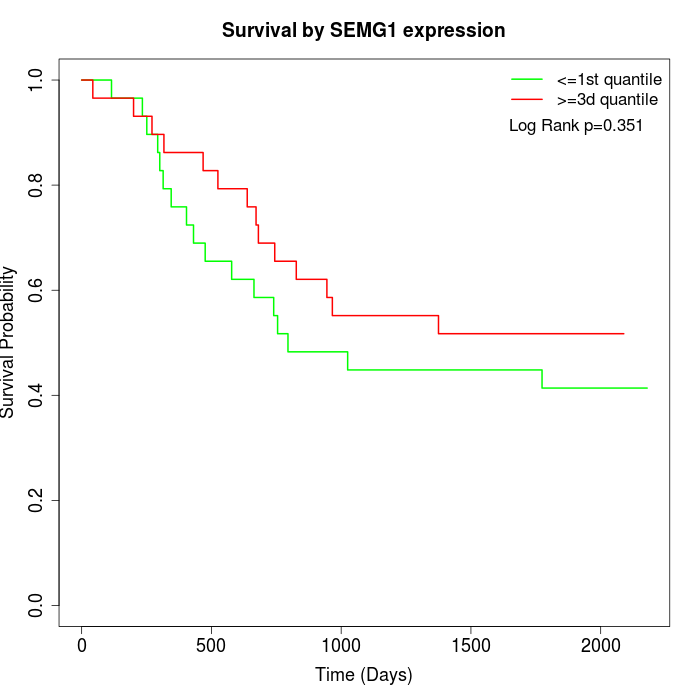
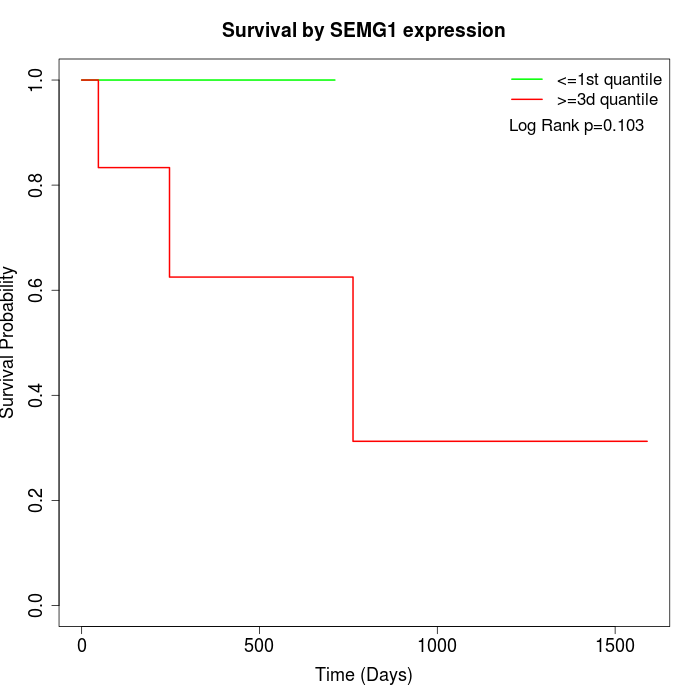
Survival by SEMG1 expression:
Note: Click image to view full size file.
Copy number change of SEMG1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SEMG1 | 6406 | 13 | 1 | 16 | |
| GSE20123 | SEMG1 | 6406 | 13 | 1 | 16 | |
| GSE43470 | SEMG1 | 6406 | 12 | 1 | 30 | |
| GSE46452 | SEMG1 | 6406 | 29 | 0 | 30 | |
| GSE47630 | SEMG1 | 6406 | 24 | 1 | 15 | |
| GSE54993 | SEMG1 | 6406 | 0 | 17 | 53 | |
| GSE54994 | SEMG1 | 6406 | 25 | 0 | 28 | |
| GSE60625 | SEMG1 | 6406 | 0 | 0 | 11 | |
| GSE74703 | SEMG1 | 6406 | 10 | 1 | 25 | |
| GSE74704 | SEMG1 | 6406 | 9 | 0 | 11 | |
| TCGA | SEMG1 | 6406 | 45 | 3 | 48 |
Total number of gains: 180; Total number of losses: 25; Total Number of normals: 283.
Somatic mutations of SEMG1:
Generating mutation plots.
Highly correlated genes for SEMG1:
Showing top 20/193 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SEMG1 | NMNAT2 | 0.683043 | 3 | 0 | 3 |
| SEMG1 | OR7C1 | 0.677616 | 4 | 0 | 3 |
| SEMG1 | BPY2 | 0.670834 | 3 | 0 | 3 |
| SEMG1 | LINC01197 | 0.665433 | 3 | 0 | 3 |
| SEMG1 | C14orf132 | 0.662741 | 3 | 0 | 3 |
| SEMG1 | KCNC4 | 0.655013 | 3 | 0 | 3 |
| SEMG1 | SPX | 0.639129 | 4 | 0 | 3 |
| SEMG1 | CHRNA9 | 0.638404 | 3 | 0 | 3 |
| SEMG1 | AFF3 | 0.637175 | 3 | 0 | 3 |
| SEMG1 | CHRND | 0.636493 | 3 | 0 | 3 |
| SEMG1 | GPR135 | 0.632627 | 3 | 0 | 3 |
| SEMG1 | EPS8L3 | 0.62419 | 4 | 0 | 4 |
| SEMG1 | PHLPP2 | 0.621319 | 4 | 0 | 3 |
| SEMG1 | PSG1 | 0.616233 | 4 | 0 | 3 |
| SEMG1 | CTRC | 0.615706 | 4 | 0 | 3 |
| SEMG1 | TMEM31 | 0.615101 | 3 | 0 | 3 |
| SEMG1 | CASP10 | 0.611135 | 3 | 0 | 3 |
| SEMG1 | NPY5R | 0.61042 | 4 | 0 | 3 |
| SEMG1 | CALCA | 0.604107 | 4 | 0 | 3 |
| SEMG1 | ZNF835 | 0.601666 | 3 | 0 | 3 |
For details and further investigation, click here