| Full name: cholinergic receptor nicotinic alpha 9 subunit | Alias Symbol: NACHRA9 | ||
| Type: protein-coding gene | Cytoband: 4p14 | ||
| Entrez ID: 55584 | HGNC ID: HGNC:14079 | Ensembl Gene: ENSG00000174343 | OMIM ID: 605116 |
| Drug and gene relationship at DGIdb | |||
Expression of CHRNA9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CHRNA9 | 55584 | 221107_at | 0.5367 | 0.4613 | |
| GSE20347 | CHRNA9 | 55584 | 221107_at | 0.1527 | 0.2910 | |
| GSE23400 | CHRNA9 | 55584 | 221107_at | 0.0035 | 0.9676 | |
| GSE26886 | CHRNA9 | 55584 | 221107_at | 0.1317 | 0.1957 | |
| GSE29001 | CHRNA9 | 55584 | 221107_at | -0.0395 | 0.8835 | |
| GSE38129 | CHRNA9 | 55584 | 221107_at | 0.0230 | 0.8582 | |
| GSE45670 | CHRNA9 | 55584 | 221107_at | 0.6041 | 0.2091 | |
| GSE63941 | CHRNA9 | 55584 | 221107_at | 0.8322 | 0.5659 | |
| GSE77861 | CHRNA9 | 55584 | 221107_at | -0.1527 | 0.0783 | |
| TCGA | CHRNA9 | 55584 | RNAseq | 4.9688 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 0.
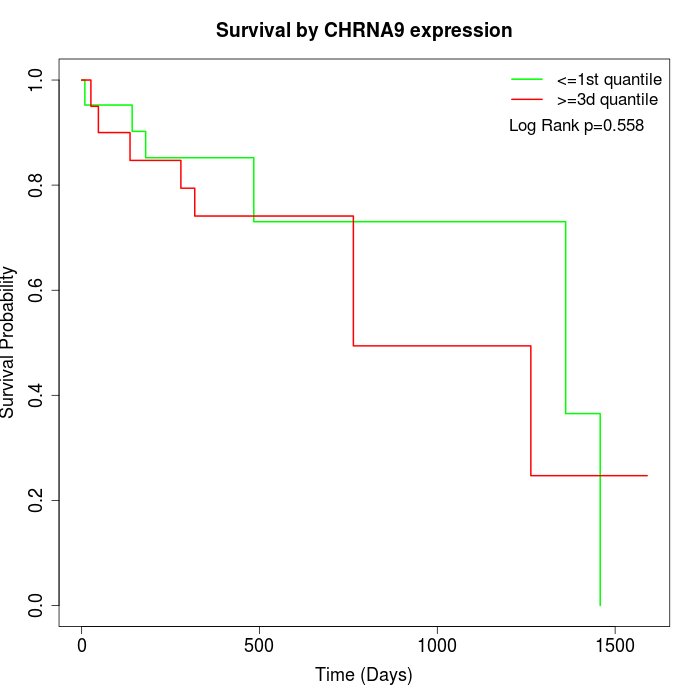
Survival by CHRNA9 expression:
Note: Click image to view full size file.
Copy number change of CHRNA9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CHRNA9 | 55584 | 0 | 16 | 14 | |
| GSE20123 | CHRNA9 | 55584 | 0 | 16 | 14 | |
| GSE43470 | CHRNA9 | 55584 | 0 | 15 | 28 | |
| GSE46452 | CHRNA9 | 55584 | 1 | 36 | 22 | |
| GSE47630 | CHRNA9 | 55584 | 1 | 19 | 20 | |
| GSE54993 | CHRNA9 | 55584 | 7 | 0 | 63 | |
| GSE54994 | CHRNA9 | 55584 | 3 | 10 | 40 | |
| GSE60625 | CHRNA9 | 55584 | 0 | 0 | 11 | |
| GSE74703 | CHRNA9 | 55584 | 0 | 12 | 24 | |
| GSE74704 | CHRNA9 | 55584 | 0 | 9 | 11 | |
| TCGA | CHRNA9 | 55584 | 10 | 45 | 41 |
Total number of gains: 22; Total number of losses: 178; Total Number of normals: 288.
Somatic mutations of CHRNA9:
Generating mutation plots.
Highly correlated genes for CHRNA9:
Showing top 20/83 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CHRNA9 | CRYGC | 0.672401 | 4 | 0 | 3 |
| CHRNA9 | SH2D5 | 0.668904 | 3 | 0 | 3 |
| CHRNA9 | DCX | 0.668699 | 3 | 0 | 3 |
| CHRNA9 | CNTNAP5 | 0.667817 | 3 | 0 | 3 |
| CHRNA9 | IL22 | 0.663876 | 3 | 0 | 3 |
| CHRNA9 | FOXP1 | 0.647312 | 3 | 0 | 3 |
| CHRNA9 | NES | 0.645596 | 3 | 0 | 3 |
| CHRNA9 | BSN | 0.640915 | 4 | 0 | 4 |
| CHRNA9 | SEMG1 | 0.638404 | 3 | 0 | 3 |
| CHRNA9 | PDE6C | 0.63557 | 3 | 0 | 3 |
| CHRNA9 | LIN28A | 0.633003 | 4 | 0 | 3 |
| CHRNA9 | LIM2 | 0.631805 | 3 | 0 | 3 |
| CHRNA9 | HOXB6 | 0.6205 | 3 | 0 | 3 |
| CHRNA9 | CNGA3 | 0.619036 | 3 | 0 | 3 |
| CHRNA9 | GLTPD2 | 0.617489 | 3 | 0 | 3 |
| CHRNA9 | TRIM10 | 0.616754 | 4 | 0 | 3 |
| CHRNA9 | KCNJ1 | 0.616365 | 4 | 0 | 3 |
| CHRNA9 | HYAL4 | 0.610355 | 4 | 0 | 3 |
| CHRNA9 | SPINT3 | 0.609295 | 3 | 0 | 3 |
| CHRNA9 | DLX1 | 0.603437 | 3 | 0 | 3 |
For details and further investigation, click here