| Full name: nicotinamide nucleotide adenylyltransferase 2 | Alias Symbol: KIAA0479|PNAT2 | ||
| Type: protein-coding gene | Cytoband: 1q25.3 | ||
| Entrez ID: 23057 | HGNC ID: HGNC:16789 | Ensembl Gene: ENSG00000157064 | OMIM ID: 608701 |
| Drug and gene relationship at DGIdb | |||
Expression of NMNAT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NMNAT2 | 23057 | 209755_at | 0.0336 | 0.9537 | |
| GSE20347 | NMNAT2 | 23057 | 209755_at | -0.0535 | 0.5426 | |
| GSE23400 | NMNAT2 | 23057 | 209755_at | -0.2285 | 0.0000 | |
| GSE26886 | NMNAT2 | 23057 | 209755_at | -0.1144 | 0.4307 | |
| GSE29001 | NMNAT2 | 23057 | 209755_at | -0.1096 | 0.3967 | |
| GSE38129 | NMNAT2 | 23057 | 209755_at | -0.2976 | 0.0204 | |
| GSE45670 | NMNAT2 | 23057 | 1552712_a_at | 0.2101 | 0.1332 | |
| GSE53622 | NMNAT2 | 23057 | 9428 | -0.1479 | 0.2131 | |
| GSE53624 | NMNAT2 | 23057 | 9428 | -0.0456 | 0.7595 | |
| GSE63941 | NMNAT2 | 23057 | 1552712_a_at | 0.1798 | 0.7668 | |
| GSE77861 | NMNAT2 | 23057 | 1552712_a_at | 0.0337 | 0.7787 | |
| GSE97050 | NMNAT2 | 23057 | A_23_P354908 | -0.0792 | 0.7798 | |
| SRP064894 | NMNAT2 | 23057 | RNAseq | 0.6958 | 0.0153 | |
| SRP133303 | NMNAT2 | 23057 | RNAseq | 0.9797 | 0.0179 | |
| SRP159526 | NMNAT2 | 23057 | RNAseq | 0.2969 | 0.6189 | |
| SRP193095 | NMNAT2 | 23057 | RNAseq | 1.9640 | 0.0000 | |
| SRP219564 | NMNAT2 | 23057 | RNAseq | 0.8901 | 0.2300 | |
| TCGA | NMNAT2 | 23057 | RNAseq | -0.1468 | 0.5033 |
Upregulated datasets: 1; Downregulated datasets: 0.
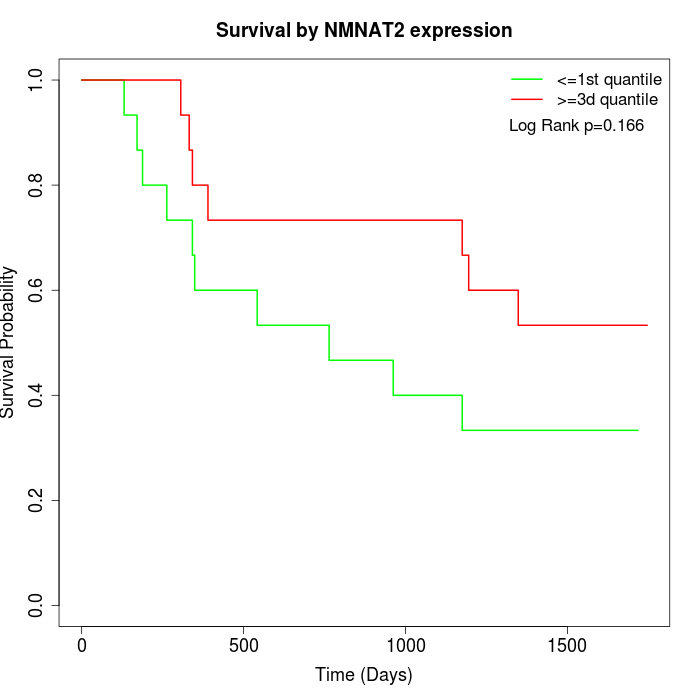
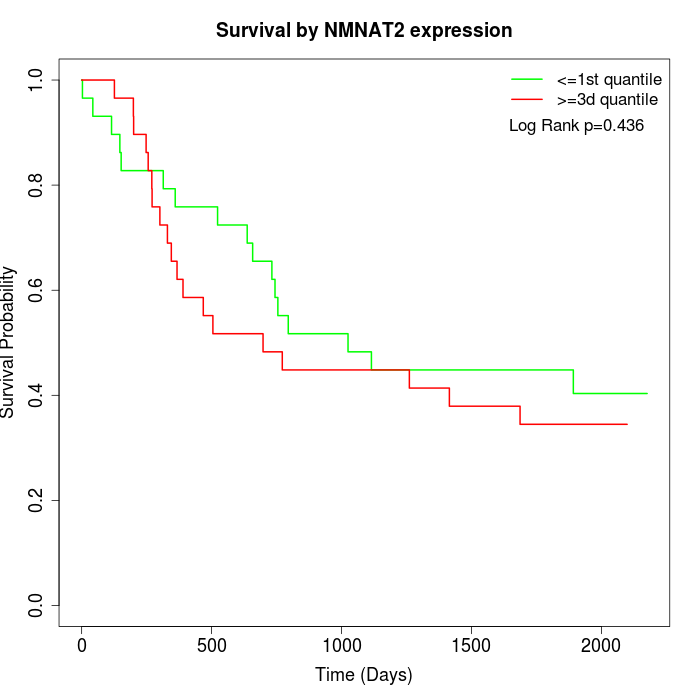
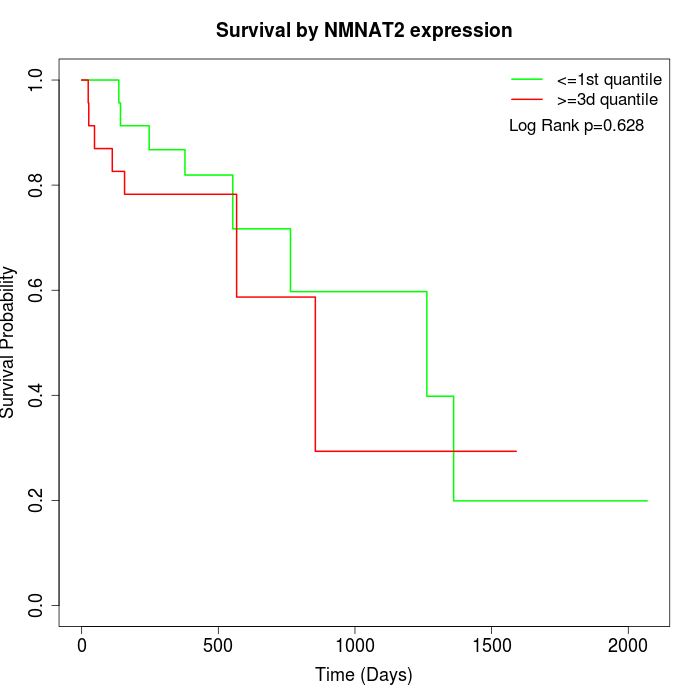
Survival by NMNAT2 expression:
Note: Click image to view full size file.
Copy number change of NMNAT2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NMNAT2 | 23057 | 12 | 0 | 18 | |
| GSE20123 | NMNAT2 | 23057 | 12 | 0 | 18 | |
| GSE43470 | NMNAT2 | 23057 | 7 | 1 | 35 | |
| GSE46452 | NMNAT2 | 23057 | 3 | 1 | 55 | |
| GSE47630 | NMNAT2 | 23057 | 14 | 0 | 26 | |
| GSE54993 | NMNAT2 | 23057 | 0 | 6 | 64 | |
| GSE54994 | NMNAT2 | 23057 | 15 | 0 | 38 | |
| GSE60625 | NMNAT2 | 23057 | 0 | 0 | 11 | |
| GSE74703 | NMNAT2 | 23057 | 7 | 1 | 28 | |
| GSE74704 | NMNAT2 | 23057 | 5 | 0 | 15 | |
| TCGA | NMNAT2 | 23057 | 40 | 3 | 53 |
Total number of gains: 115; Total number of losses: 12; Total Number of normals: 361.
Somatic mutations of NMNAT2:
Generating mutation plots.
Highly correlated genes for NMNAT2:
Showing top 20/623 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NMNAT2 | PCGF2 | 0.778926 | 3 | 0 | 3 |
| NMNAT2 | CCDC144A | 0.769855 | 3 | 0 | 3 |
| NMNAT2 | INO80D | 0.749508 | 4 | 0 | 4 |
| NMNAT2 | CER1 | 0.748754 | 3 | 0 | 3 |
| NMNAT2 | P2RX6 | 0.737546 | 3 | 0 | 3 |
| NMNAT2 | OR10J1 | 0.73623 | 3 | 0 | 3 |
| NMNAT2 | SYDE1 | 0.734755 | 4 | 0 | 4 |
| NMNAT2 | KRT38 | 0.727817 | 3 | 0 | 3 |
| NMNAT2 | C1orf68 | 0.726451 | 3 | 0 | 3 |
| NMNAT2 | NPY1R | 0.726078 | 3 | 0 | 3 |
| NMNAT2 | GABRR2 | 0.723759 | 4 | 0 | 4 |
| NMNAT2 | TM4SF18 | 0.712469 | 4 | 0 | 3 |
| NMNAT2 | CETN1 | 0.710363 | 3 | 0 | 3 |
| NMNAT2 | MRPL41 | 0.709028 | 4 | 0 | 4 |
| NMNAT2 | NLGN4X | 0.707233 | 3 | 0 | 3 |
| NMNAT2 | GNAT2 | 0.707138 | 5 | 0 | 4 |
| NMNAT2 | IFNA16 | 0.705674 | 3 | 0 | 3 |
| NMNAT2 | NES | 0.705435 | 4 | 0 | 3 |
| NMNAT2 | GDF10 | 0.704674 | 3 | 0 | 3 |
| NMNAT2 | MCAM | 0.704416 | 3 | 0 | 3 |
For details and further investigation, click here