| Full name: serum/glucocorticoid regulated kinase 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 6q23.2 | ||
| Entrez ID: 6446 | HGNC ID: HGNC:10810 | Ensembl Gene: ENSG00000118515 | OMIM ID: 602958 |
| Drug and gene relationship at DGIdb | |||
SGK1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04068 | FoxO signaling pathway | |
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa04960 | Aldosterone-regulated sodium reabsorption |
Expression of SGK1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SGK1 | 6446 | 201739_at | 0.5113 | 0.2535 | |
| GSE20347 | SGK1 | 6446 | 201739_at | 0.3902 | 0.2495 | |
| GSE23400 | SGK1 | 6446 | 201739_at | 0.3472 | 0.0251 | |
| GSE26886 | SGK1 | 6446 | 201739_at | -0.0218 | 0.9567 | |
| GSE29001 | SGK1 | 6446 | 201739_at | 0.1227 | 0.6571 | |
| GSE38129 | SGK1 | 6446 | 201739_at | 0.6784 | 0.0486 | |
| GSE45670 | SGK1 | 6446 | 201739_at | 0.7656 | 0.0019 | |
| GSE53622 | SGK1 | 6446 | 36650 | 0.4137 | 0.0024 | |
| GSE53624 | SGK1 | 6446 | 36650 | 0.2603 | 0.0046 | |
| GSE63941 | SGK1 | 6446 | 201739_at | -2.1633 | 0.0926 | |
| GSE77861 | SGK1 | 6446 | 201739_at | 0.6814 | 0.2527 | |
| GSE97050 | SGK1 | 6446 | A_23_P19673 | 0.7065 | 0.1205 | |
| SRP007169 | SGK1 | 6446 | RNAseq | 0.1679 | 0.6833 | |
| SRP008496 | SGK1 | 6446 | RNAseq | 0.3013 | 0.4242 | |
| SRP064894 | SGK1 | 6446 | RNAseq | 0.5843 | 0.0465 | |
| SRP133303 | SGK1 | 6446 | RNAseq | 1.0307 | 0.0028 | |
| SRP159526 | SGK1 | 6446 | RNAseq | 0.3521 | 0.5259 | |
| SRP193095 | SGK1 | 6446 | RNAseq | 0.7913 | 0.0005 | |
| SRP219564 | SGK1 | 6446 | RNAseq | 0.4136 | 0.3858 | |
| TCGA | SGK1 | 6446 | RNAseq | -0.0506 | 0.5411 |
Upregulated datasets: 1; Downregulated datasets: 0.
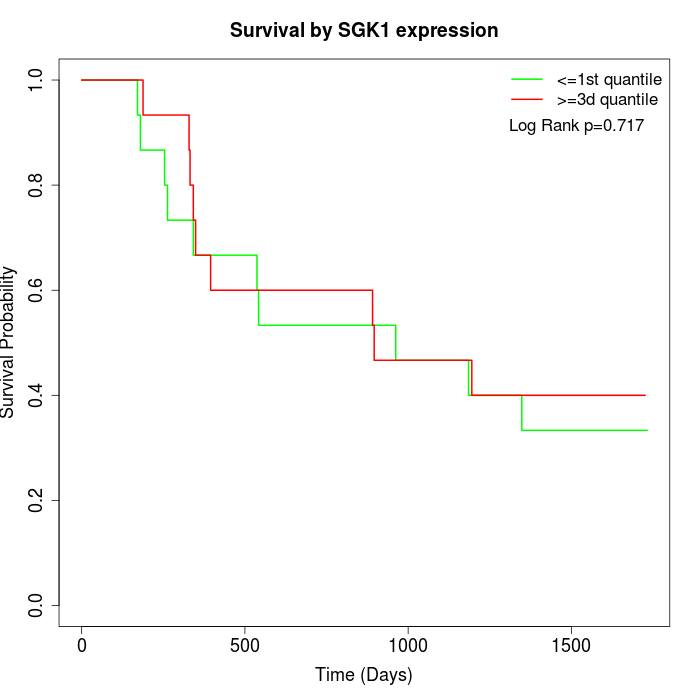
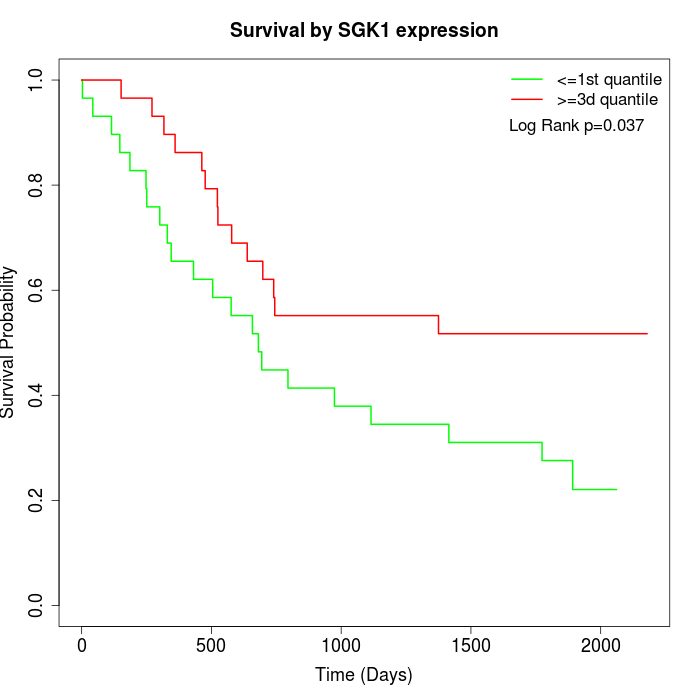
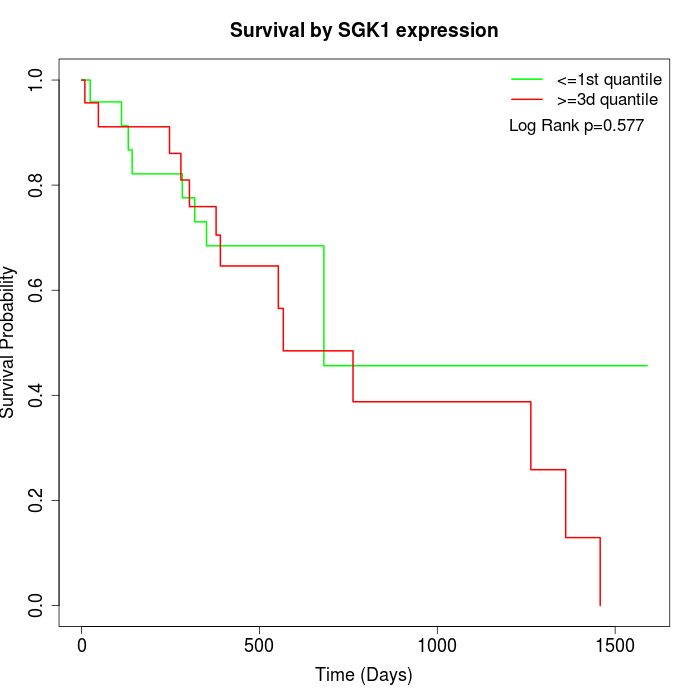
Survival by SGK1 expression:
Note: Click image to view full size file.
Copy number change of SGK1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SGK1 | 6446 | 4 | 4 | 22 | |
| GSE20123 | SGK1 | 6446 | 4 | 3 | 23 | |
| GSE43470 | SGK1 | 6446 | 5 | 0 | 38 | |
| GSE46452 | SGK1 | 6446 | 3 | 10 | 46 | |
| GSE47630 | SGK1 | 6446 | 10 | 4 | 26 | |
| GSE54993 | SGK1 | 6446 | 3 | 2 | 65 | |
| GSE54994 | SGK1 | 6446 | 10 | 7 | 36 | |
| GSE60625 | SGK1 | 6446 | 0 | 1 | 10 | |
| GSE74703 | SGK1 | 6446 | 4 | 0 | 32 | |
| GSE74704 | SGK1 | 6446 | 1 | 1 | 18 | |
| TCGA | SGK1 | 6446 | 12 | 18 | 66 |
Total number of gains: 56; Total number of losses: 50; Total Number of normals: 382.
Somatic mutations of SGK1:
Generating mutation plots.
Highly correlated genes for SGK1:
Showing top 20/121 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SGK1 | ZDHHC9 | 0.674086 | 3 | 0 | 3 |
| SGK1 | CD38 | 0.654535 | 3 | 0 | 3 |
| SGK1 | MRPL4 | 0.64379 | 3 | 0 | 3 |
| SGK1 | ABRACL | 0.637523 | 4 | 0 | 4 |
| SGK1 | CEP350 | 0.628373 | 3 | 0 | 3 |
| SGK1 | GATAD2A | 0.627979 | 4 | 0 | 4 |
| SGK1 | FBXO41 | 0.622981 | 4 | 0 | 3 |
| SGK1 | HPCAL1 | 0.621725 | 3 | 0 | 3 |
| SGK1 | CMIP | 0.613634 | 4 | 0 | 3 |
| SGK1 | GAA | 0.608008 | 3 | 0 | 3 |
| SGK1 | DNAJB11 | 0.60305 | 5 | 0 | 3 |
| SGK1 | CXXC1 | 0.600094 | 3 | 0 | 3 |
| SGK1 | FAM49B | 0.599259 | 4 | 0 | 3 |
| SGK1 | EPHB1 | 0.596971 | 5 | 0 | 4 |
| SGK1 | ABL2 | 0.596001 | 3 | 0 | 3 |
| SGK1 | CORO1A | 0.595301 | 3 | 0 | 3 |
| SGK1 | COMMD5 | 0.594833 | 3 | 0 | 3 |
| SGK1 | MYPOP | 0.591483 | 4 | 0 | 3 |
| SGK1 | RAB3IP | 0.590877 | 5 | 0 | 3 |
| SGK1 | API5 | 0.590147 | 4 | 0 | 4 |
For details and further investigation, click here