| Full name: ABL proto-oncogene 2, non-receptor tyrosine kinase | Alias Symbol: ARG | ||
| Type: protein-coding gene | Cytoband: 1q25.2 | ||
| Entrez ID: 27 | HGNC ID: HGNC:77 | Ensembl Gene: ENSG00000143322 | OMIM ID: 164690 |
| Drug and gene relationship at DGIdb | |||
ABL2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04012 | ErbB signaling pathway | |
| hsa04014 | Ras signaling pathway |
Expression of ABL2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ABL2 | 27 | 226893_at | 1.0456 | 0.0102 | |
| GSE20347 | ABL2 | 27 | 206411_s_at | 0.4519 | 0.0015 | |
| GSE23400 | ABL2 | 27 | 206411_s_at | 0.1407 | 0.0060 | |
| GSE26886 | ABL2 | 27 | 226893_at | 1.5126 | 0.0000 | |
| GSE29001 | ABL2 | 27 | 206411_s_at | 0.0344 | 0.8227 | |
| GSE38129 | ABL2 | 27 | 206411_s_at | 0.3638 | 0.0047 | |
| GSE45670 | ABL2 | 27 | 231907_at | 0.4160 | 0.1695 | |
| GSE53622 | ABL2 | 27 | 58248 | 0.8833 | 0.0000 | |
| GSE53624 | ABL2 | 27 | 58248 | 1.3901 | 0.0000 | |
| GSE63941 | ABL2 | 27 | 226893_at | -1.6835 | 0.0013 | |
| GSE77861 | ABL2 | 27 | 226893_at | 0.8390 | 0.0024 | |
| GSE97050 | ABL2 | 27 | A_33_P3413671 | 0.9967 | 0.0631 | |
| SRP007169 | ABL2 | 27 | RNAseq | 1.9803 | 0.0000 | |
| SRP008496 | ABL2 | 27 | RNAseq | 2.4475 | 0.0000 | |
| SRP064894 | ABL2 | 27 | RNAseq | 1.2298 | 0.0000 | |
| SRP133303 | ABL2 | 27 | RNAseq | 1.3905 | 0.0000 | |
| SRP159526 | ABL2 | 27 | RNAseq | 1.0623 | 0.0004 | |
| SRP193095 | ABL2 | 27 | RNAseq | 1.8417 | 0.0000 | |
| SRP219564 | ABL2 | 27 | RNAseq | 1.5854 | 0.0022 | |
| TCGA | ABL2 | 27 | RNAseq | 0.1034 | 0.0685 |
Upregulated datasets: 10; Downregulated datasets: 1.
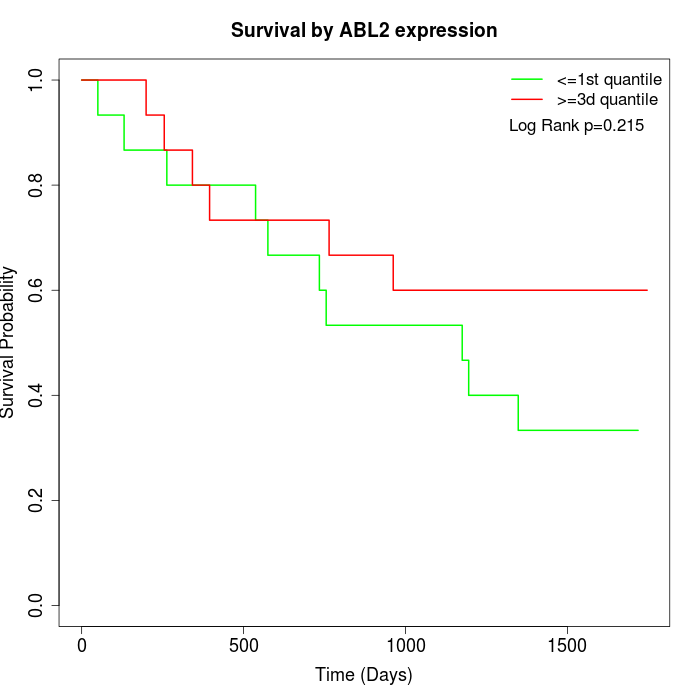
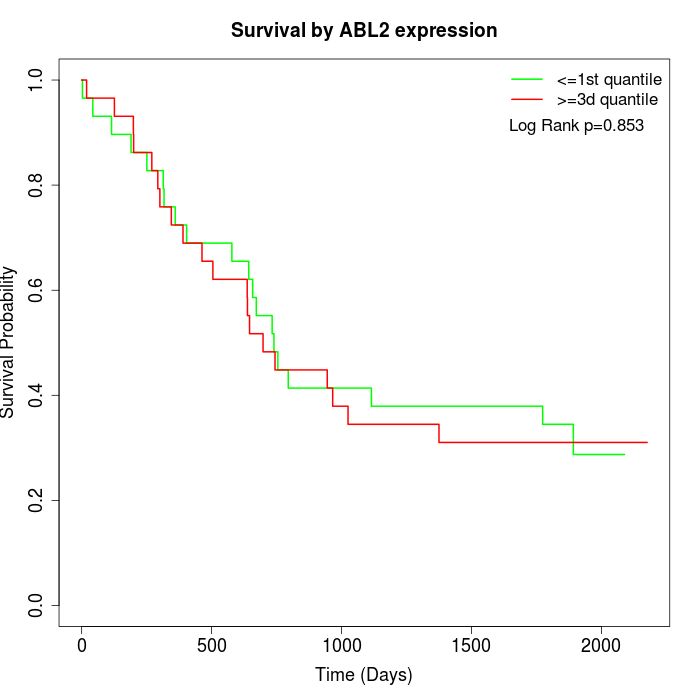
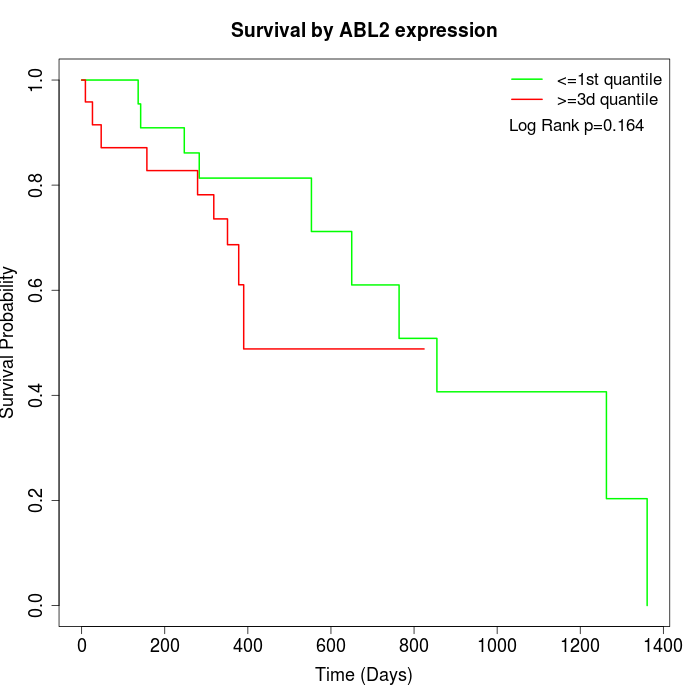
Survival by ABL2 expression:
Note: Click image to view full size file.
Copy number change of ABL2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ABL2 | 27 | 12 | 0 | 18 | |
| GSE20123 | ABL2 | 27 | 12 | 0 | 18 | |
| GSE43470 | ABL2 | 27 | 9 | 1 | 33 | |
| GSE46452 | ABL2 | 27 | 3 | 1 | 55 | |
| GSE47630 | ABL2 | 27 | 14 | 0 | 26 | |
| GSE54993 | ABL2 | 27 | 0 | 6 | 64 | |
| GSE54994 | ABL2 | 27 | 15 | 0 | 38 | |
| GSE60625 | ABL2 | 27 | 0 | 0 | 11 | |
| GSE74703 | ABL2 | 27 | 8 | 1 | 27 | |
| GSE74704 | ABL2 | 27 | 5 | 0 | 15 | |
| TCGA | ABL2 | 27 | 40 | 3 | 53 |
Total number of gains: 118; Total number of losses: 12; Total Number of normals: 358.
Somatic mutations of ABL2:
Generating mutation plots.
Highly correlated genes for ABL2:
Showing top 20/1517 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ABL2 | CD200 | 0.783046 | 3 | 0 | 3 |
| ABL2 | PLEKHM3 | 0.78257 | 3 | 0 | 3 |
| ABL2 | FADS1 | 0.771272 | 3 | 0 | 3 |
| ABL2 | FAM89A | 0.769532 | 3 | 0 | 3 |
| ABL2 | RBM15B | 0.766838 | 3 | 0 | 3 |
| ABL2 | FCGR3A | 0.763722 | 3 | 0 | 3 |
| ABL2 | KTI12 | 0.760389 | 3 | 0 | 3 |
| ABL2 | SLC41A1 | 0.75845 | 8 | 0 | 8 |
| ABL2 | RAB34 | 0.755839 | 3 | 0 | 3 |
| ABL2 | ZNF281 | 0.753849 | 10 | 0 | 10 |
| ABL2 | PYCR2 | 0.748702 | 3 | 0 | 3 |
| ABL2 | SH3RF3 | 0.746262 | 4 | 0 | 4 |
| ABL2 | FMNL2 | 0.742914 | 8 | 0 | 8 |
| ABL2 | TK2 | 0.742753 | 3 | 0 | 3 |
| ABL2 | PTPN4 | 0.741924 | 4 | 0 | 4 |
| ABL2 | GGNBP2 | 0.741546 | 4 | 0 | 4 |
| ABL2 | XPR1 | 0.739208 | 7 | 0 | 7 |
| ABL2 | TRIB1 | 0.736467 | 3 | 0 | 3 |
| ABL2 | TMEM263 | 0.736056 | 6 | 0 | 6 |
| ABL2 | SLC39A3 | 0.73518 | 4 | 0 | 4 |
For details and further investigation, click here