| Full name: solute carrier family 22 member 16 | Alias Symbol: FLIPT2|CT2|OKB1|OAT6 | ||
| Type: protein-coding gene | Cytoband: 6q21 | ||
| Entrez ID: 85413 | HGNC ID: HGNC:20302 | Ensembl Gene: ENSG00000004809 | OMIM ID: 608276 |
| Drug and gene relationship at DGIdb | |||
Expression of SLC22A16:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SLC22A16 | 85413 | 232232_s_at | 0.2834 | 0.5183 | |
| GSE26886 | SLC22A16 | 85413 | 232232_s_at | -0.1242 | 0.3570 | |
| GSE45670 | SLC22A16 | 85413 | 232232_s_at | 0.1056 | 0.2422 | |
| GSE53622 | SLC22A16 | 85413 | 95879 | 0.2300 | 0.0569 | |
| GSE53624 | SLC22A16 | 85413 | 95879 | 1.2274 | 0.0000 | |
| GSE63941 | SLC22A16 | 85413 | 232232_s_at | -0.0088 | 0.9624 | |
| GSE77861 | SLC22A16 | 85413 | 232232_s_at | -0.1136 | 0.4152 | |
| GSE97050 | SLC22A16 | 85413 | A_23_P42004 | 0.2248 | 0.2801 | |
| TCGA | SLC22A16 | 85413 | RNAseq | 1.3157 | 0.1123 |
Upregulated datasets: 1; Downregulated datasets: 0.
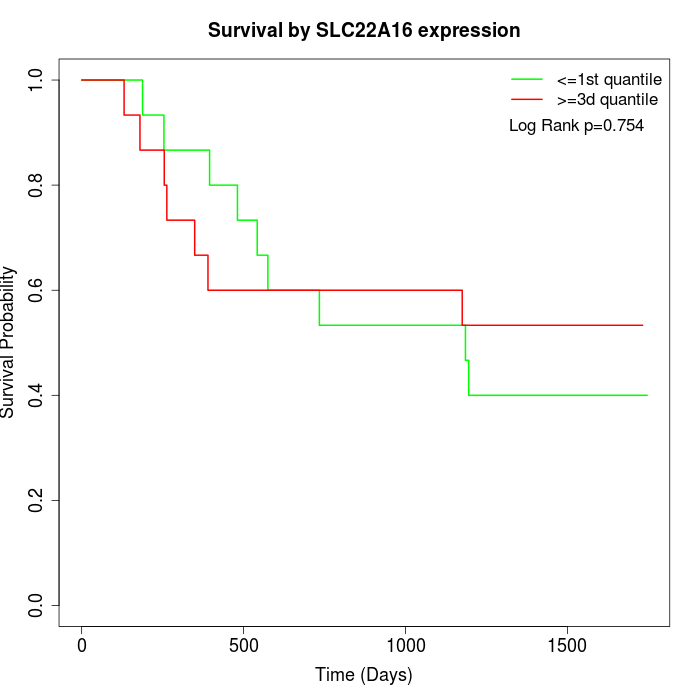
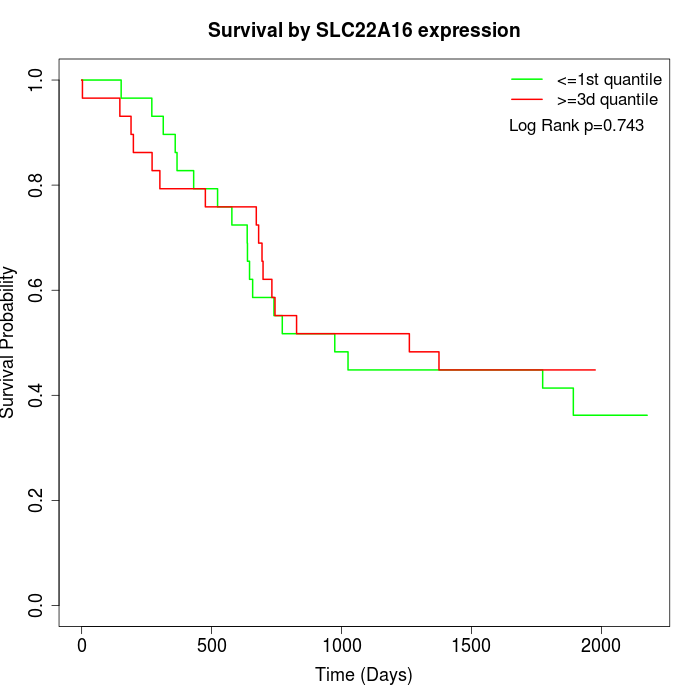
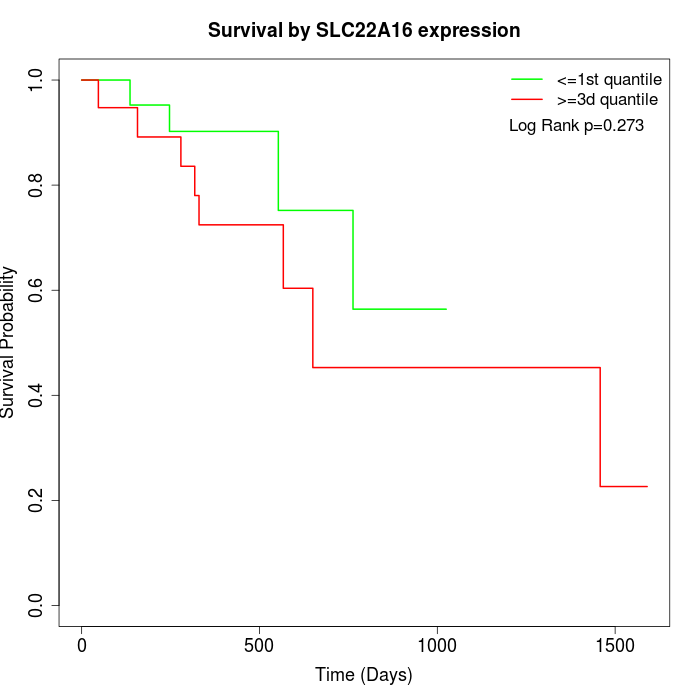
Survival by SLC22A16 expression:
Note: Click image to view full size file.
Copy number change of SLC22A16:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SLC22A16 | 85413 | 2 | 3 | 25 | |
| GSE20123 | SLC22A16 | 85413 | 3 | 2 | 25 | |
| GSE43470 | SLC22A16 | 85413 | 3 | 1 | 39 | |
| GSE46452 | SLC22A16 | 85413 | 2 | 11 | 46 | |
| GSE47630 | SLC22A16 | 85413 | 9 | 4 | 27 | |
| GSE54993 | SLC22A16 | 85413 | 3 | 2 | 65 | |
| GSE54994 | SLC22A16 | 85413 | 8 | 8 | 37 | |
| GSE60625 | SLC22A16 | 85413 | 0 | 2 | 9 | |
| GSE74703 | SLC22A16 | 85413 | 2 | 1 | 33 | |
| GSE74704 | SLC22A16 | 85413 | 1 | 0 | 19 | |
| TCGA | SLC22A16 | 85413 | 6 | 23 | 67 |
Total number of gains: 39; Total number of losses: 57; Total Number of normals: 392.
Somatic mutations of SLC22A16:
Generating mutation plots.
Highly correlated genes for SLC22A16:
Showing top 20/56 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SLC22A16 | ITGA2B | 0.776735 | 3 | 0 | 3 |
| SLC22A16 | RSPH1 | 0.776255 | 3 | 0 | 3 |
| SLC22A16 | BTNL9 | 0.706688 | 3 | 0 | 3 |
| SLC22A16 | ATP7B | 0.693869 | 4 | 0 | 3 |
| SLC22A16 | RNF157 | 0.690083 | 4 | 0 | 3 |
| SLC22A16 | AGPAT1 | 0.677514 | 3 | 0 | 3 |
| SLC22A16 | CELA3B | 0.675676 | 3 | 0 | 3 |
| SLC22A16 | POC1A | 0.675519 | 3 | 0 | 3 |
| SLC22A16 | LRFN4 | 0.671589 | 4 | 0 | 3 |
| SLC22A16 | RAD51 | 0.668102 | 3 | 0 | 3 |
| SLC22A16 | SNORA71A | 0.667753 | 4 | 0 | 3 |
| SLC22A16 | SIRPD | 0.666542 | 4 | 0 | 3 |
| SLC22A16 | SMCP | 0.642584 | 4 | 0 | 4 |
| SLC22A16 | RENBP | 0.628723 | 4 | 0 | 3 |
| SLC22A16 | CRLF2 | 0.626375 | 3 | 0 | 3 |
| SLC22A16 | TMEM235 | 0.621386 | 4 | 0 | 3 |
| SLC22A16 | DBF4B | 0.620289 | 4 | 0 | 3 |
| SLC22A16 | CCL25 | 0.614298 | 5 | 0 | 4 |
| SLC22A16 | RAB39A | 0.608929 | 3 | 0 | 3 |
| SLC22A16 | CD300LB | 0.608815 | 3 | 0 | 3 |
For details and further investigation, click here