| Full name: SMAD1 antisense RNA 2 | Alias Symbol: | ||
| Type: non-coding RNA | Cytoband: 4q31.21 | ||
| Entrez ID: 101927659 | HGNC ID: HGNC:49381 | Ensembl Gene: ENSG00000250582 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of SMAD1-AS2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SMAD1-AS2 | 101927659 | 244446_at | 0.0822 | 0.7270 | |
| GSE26886 | SMAD1-AS2 | 101927659 | 244446_at | 0.1144 | 0.2348 | |
| GSE45670 | SMAD1-AS2 | 101927659 | 244446_at | 0.0371 | 0.6808 | |
| GSE53622 | SMAD1-AS2 | 101927659 | 70730 | 0.1901 | 0.0569 | |
| GSE53624 | SMAD1-AS2 | 101927659 | 70730 | 0.5820 | 0.0000 | |
| GSE63941 | SMAD1-AS2 | 101927659 | 244446_at | 0.1393 | 0.4383 | |
| GSE77861 | SMAD1-AS2 | 101927659 | 244446_at | -0.0651 | 0.4101 | |
| SRP133303 | SMAD1-AS2 | 101927659 | RNAseq | 0.7467 | 0.0060 |
Upregulated datasets: 0; Downregulated datasets: 0.
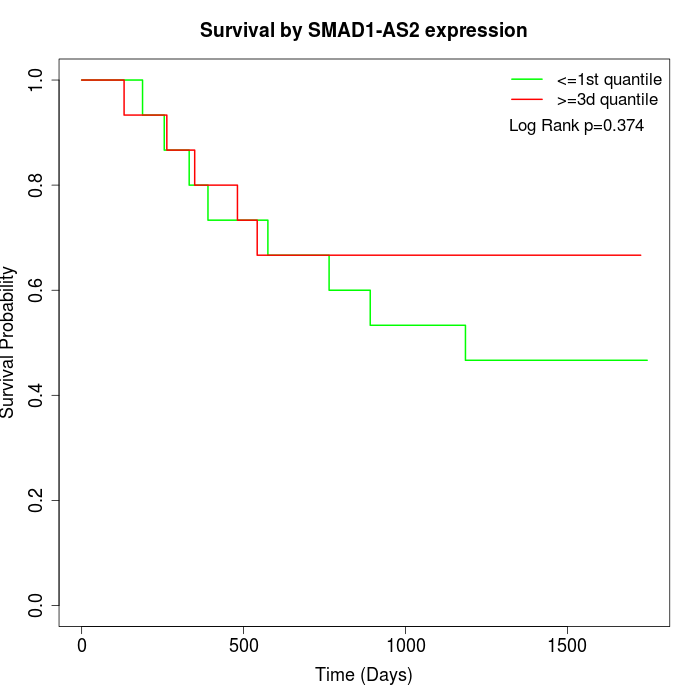
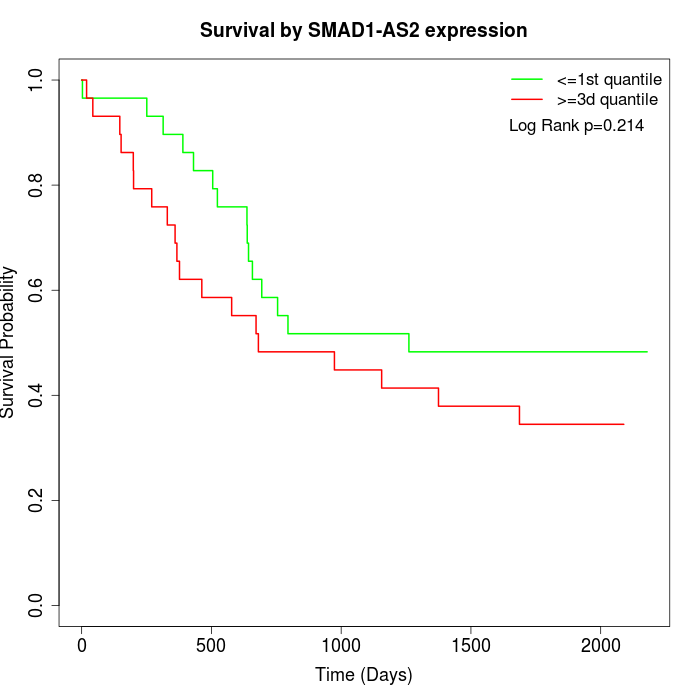
Survival by SMAD1-AS2 expression:
Note: Click image to view full size file.
Copy number change of SMAD1-AS2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SMAD1-AS2 | 101927659 | 0 | 12 | 18 | |
| GSE20123 | SMAD1-AS2 | 101927659 | 0 | 12 | 18 | |
| GSE43470 | SMAD1-AS2 | 101927659 | 0 | 13 | 30 | |
| GSE46452 | SMAD1-AS2 | 101927659 | 1 | 36 | 22 | |
| GSE47630 | SMAD1-AS2 | 101927659 | 0 | 22 | 18 | |
| GSE54993 | SMAD1-AS2 | 101927659 | 10 | 0 | 60 | |
| GSE54994 | SMAD1-AS2 | 101927659 | 2 | 11 | 40 | |
| GSE60625 | SMAD1-AS2 | 101927659 | 0 | 1 | 10 | |
| GSE74703 | SMAD1-AS2 | 101927659 | 0 | 11 | 25 | |
| GSE74704 | SMAD1-AS2 | 101927659 | 0 | 7 | 13 |
Total number of gains: 13; Total number of losses: 125; Total Number of normals: 254.
Somatic mutations of SMAD1-AS2:
Generating mutation plots.
Highly correlated genes for SMAD1-AS2:
Showing top 20/26 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SMAD1-AS2 | LIMD1-AS1 | 0.66956 | 3 | 0 | 3 |
| SMAD1-AS2 | TMEM26 | 0.617221 | 3 | 0 | 3 |
| SMAD1-AS2 | FRMD5 | 0.610467 | 3 | 0 | 3 |
| SMAD1-AS2 | LAMTOR5-AS1 | 0.592914 | 3 | 0 | 3 |
| SMAD1-AS2 | ASPDH | 0.592171 | 3 | 0 | 3 |
| SMAD1-AS2 | HECW1 | 0.591328 | 3 | 0 | 3 |
| SMAD1-AS2 | GLYCTK-AS1 | 0.58553 | 3 | 0 | 3 |
| SMAD1-AS2 | CDH7 | 0.585059 | 4 | 0 | 3 |
| SMAD1-AS2 | GATA3-AS1 | 0.581571 | 3 | 0 | 3 |
| SMAD1-AS2 | ASB11 | 0.578348 | 3 | 0 | 3 |
| SMAD1-AS2 | NFIA-AS2 | 0.57713 | 4 | 0 | 3 |
| SMAD1-AS2 | OR13C4 | 0.561026 | 4 | 0 | 3 |
| SMAD1-AS2 | OR10A3 | 0.553219 | 3 | 0 | 3 |
| SMAD1-AS2 | SLC22A11 | 0.547047 | 4 | 0 | 3 |
| SMAD1-AS2 | TTC9B | 0.538116 | 4 | 0 | 3 |
| SMAD1-AS2 | RGS8 | 0.535982 | 4 | 0 | 3 |
| SMAD1-AS2 | SLC6A13 | 0.53492 | 4 | 0 | 3 |
| SMAD1-AS2 | PERM1 | 0.527611 | 3 | 0 | 3 |
| SMAD1-AS2 | GRP | 0.52615 | 4 | 0 | 3 |
| SMAD1-AS2 | HTR1B | 0.52547 | 4 | 0 | 3 |
For details and further investigation, click here