| Full name: gastrin releasing peptide | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 18q21.32 | ||
| Entrez ID: 2922 | HGNC ID: HGNC:4605 | Ensembl Gene: ENSG00000134443 | OMIM ID: 137260 |
| Drug and gene relationship at DGIdb | |||
Expression of GRP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GRP | 2922 | 206326_at | 0.6480 | 0.3205 | |
| GSE20347 | GRP | 2922 | 206326_at | 0.6792 | 0.0039 | |
| GSE23400 | GRP | 2922 | 206326_at | 0.1497 | 0.0042 | |
| GSE26886 | GRP | 2922 | 206326_at | 1.3018 | 0.0007 | |
| GSE29001 | GRP | 2922 | 206326_at | 0.5887 | 0.0545 | |
| GSE38129 | GRP | 2922 | 206326_at | 0.8142 | 0.0327 | |
| GSE45670 | GRP | 2922 | 206326_at | 0.3686 | 0.1183 | |
| GSE53622 | GRP | 2922 | 43181 | 0.5737 | 0.0036 | |
| GSE53624 | GRP | 2922 | 43181 | 1.2753 | 0.0000 | |
| GSE63941 | GRP | 2922 | 206326_at | 0.0472 | 0.7459 | |
| GSE77861 | GRP | 2922 | 206326_at | -0.1152 | 0.1911 | |
| SRP133303 | GRP | 2922 | RNAseq | 7.1279 | 0.0000 | |
| TCGA | GRP | 2922 | RNAseq | -0.0210 | 0.9699 |
Upregulated datasets: 3; Downregulated datasets: 0.
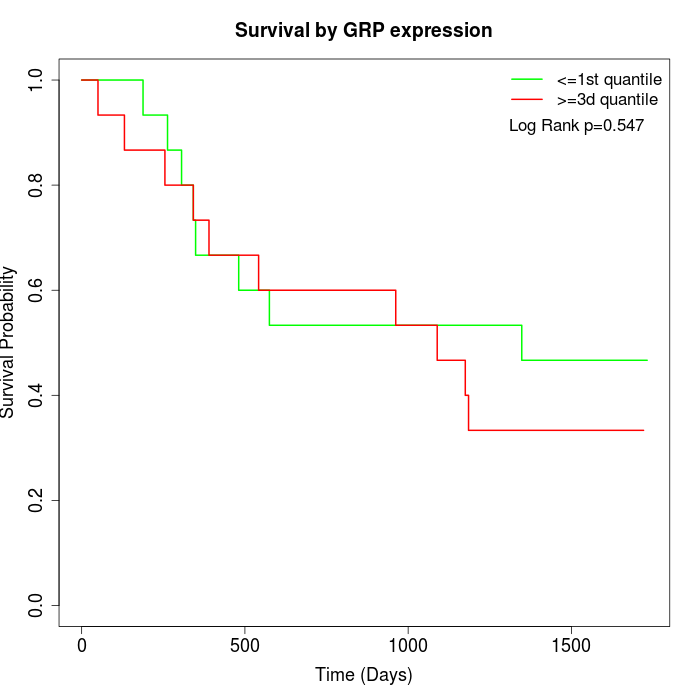
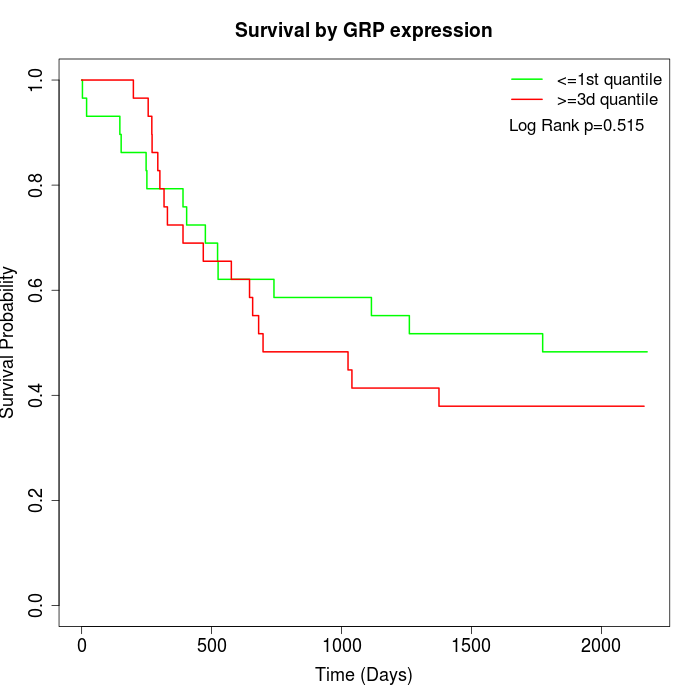
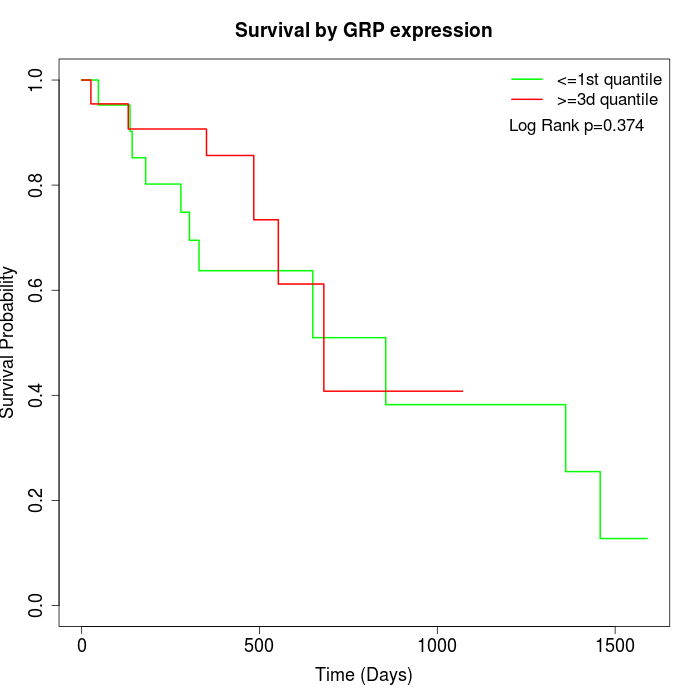
Survival by GRP expression:
Note: Click image to view full size file.
Copy number change of GRP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GRP | 2922 | 1 | 9 | 20 | |
| GSE20123 | GRP | 2922 | 1 | 9 | 20 | |
| GSE43470 | GRP | 2922 | 0 | 5 | 38 | |
| GSE46452 | GRP | 2922 | 1 | 26 | 32 | |
| GSE47630 | GRP | 2922 | 5 | 20 | 15 | |
| GSE54993 | GRP | 2922 | 10 | 0 | 60 | |
| GSE54994 | GRP | 2922 | 0 | 18 | 35 | |
| GSE60625 | GRP | 2922 | 0 | 4 | 7 | |
| GSE74703 | GRP | 2922 | 0 | 5 | 31 | |
| GSE74704 | GRP | 2922 | 1 | 6 | 13 | |
| TCGA | GRP | 2922 | 9 | 44 | 43 |
Total number of gains: 28; Total number of losses: 146; Total Number of normals: 314.
Somatic mutations of GRP:
Generating mutation plots.
Highly correlated genes for GRP:
Showing top 20/106 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GRP | CSF3 | 0.708713 | 3 | 0 | 3 |
| GRP | SYCE1L | 0.681589 | 3 | 0 | 3 |
| GRP | MCEMP1 | 0.669411 | 3 | 0 | 3 |
| GRP | ADAMTS12 | 0.663521 | 5 | 0 | 4 |
| GRP | ACD | 0.647008 | 4 | 0 | 3 |
| GRP | PDGFB | 0.641383 | 4 | 0 | 3 |
| GRP | KIAA0319 | 0.636973 | 3 | 0 | 3 |
| GRP | GRM3 | 0.627388 | 4 | 0 | 3 |
| GRP | SEZ6L2 | 0.62451 | 4 | 0 | 4 |
| GRP | COL24A1 | 0.61748 | 4 | 0 | 3 |
| GRP | ASPDH | 0.616703 | 3 | 0 | 3 |
| GRP | ICAM5 | 0.613209 | 5 | 0 | 4 |
| GRP | CA6 | 0.613074 | 4 | 0 | 3 |
| GRP | FAM71B | 0.603075 | 3 | 0 | 3 |
| GRP | DBN1 | 0.602528 | 3 | 0 | 3 |
| GRP | UBA2 | 0.600905 | 3 | 0 | 3 |
| GRP | EPHB4 | 0.589207 | 4 | 0 | 4 |
| GRP | ZNF469 | 0.589122 | 6 | 0 | 3 |
| GRP | LRP3 | 0.585986 | 4 | 0 | 3 |
| GRP | GABRR2 | 0.581948 | 3 | 0 | 3 |
For details and further investigation, click here