| Full name: SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 | Alias Symbol: BAF60C|Rsc6p|CRACD3 | ||
| Type: protein-coding gene | Cytoband: 7q36.1 | ||
| Entrez ID: 6604 | HGNC ID: HGNC:11108 | Ensembl Gene: ENSG00000082014 | OMIM ID: 601737 |
| Drug and gene relationship at DGIdb | |||
Expression of SMARCD3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SMARCD3 | 6604 | 204099_at | -0.6909 | 0.3801 | |
| GSE20347 | SMARCD3 | 6604 | 204099_at | -0.3115 | 0.0389 | |
| GSE23400 | SMARCD3 | 6604 | 204099_at | -0.3574 | 0.0001 | |
| GSE26886 | SMARCD3 | 6604 | 231144_at | 0.1760 | 0.1876 | |
| GSE29001 | SMARCD3 | 6604 | 204099_at | -0.1470 | 0.6611 | |
| GSE38129 | SMARCD3 | 6604 | 204099_at | -0.5301 | 0.0223 | |
| GSE45670 | SMARCD3 | 6604 | 204099_at | -0.4534 | 0.0397 | |
| GSE53622 | SMARCD3 | 6604 | 12739 | -0.0422 | 0.6791 | |
| GSE53624 | SMARCD3 | 6604 | 12739 | -0.2008 | 0.0045 | |
| GSE63941 | SMARCD3 | 6604 | 204099_at | -1.7177 | 0.0069 | |
| GSE77861 | SMARCD3 | 6604 | 204099_at | 0.3449 | 0.0910 | |
| GSE97050 | SMARCD3 | 6604 | A_23_P122852 | -0.5130 | 0.2211 | |
| SRP007169 | SMARCD3 | 6604 | RNAseq | 0.1332 | 0.8737 | |
| SRP064894 | SMARCD3 | 6604 | RNAseq | 0.6380 | 0.0886 | |
| SRP133303 | SMARCD3 | 6604 | RNAseq | -0.3338 | 0.1354 | |
| SRP159526 | SMARCD3 | 6604 | RNAseq | 0.2855 | 0.3795 | |
| SRP193095 | SMARCD3 | 6604 | RNAseq | 0.6109 | 0.0011 | |
| SRP219564 | SMARCD3 | 6604 | RNAseq | -0.0710 | 0.9082 | |
| TCGA | SMARCD3 | 6604 | RNAseq | -0.4681 | 0.0024 |
Upregulated datasets: 0; Downregulated datasets: 1.
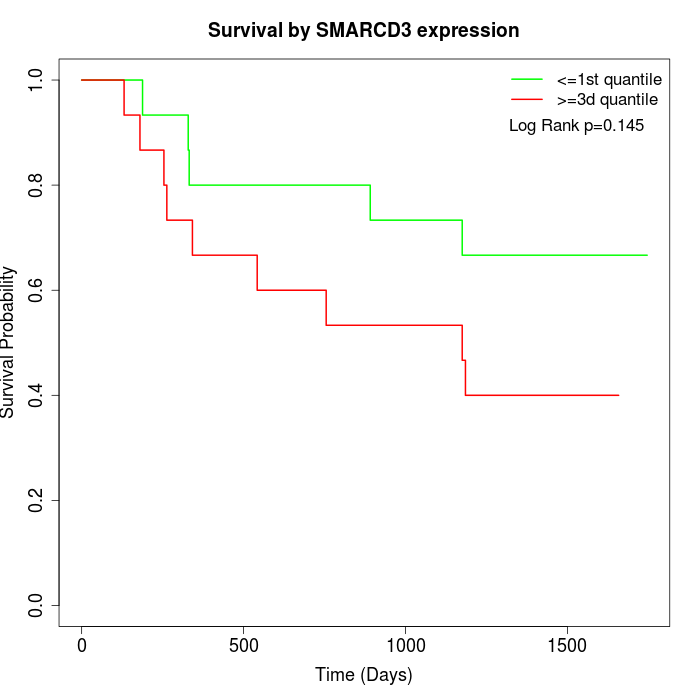
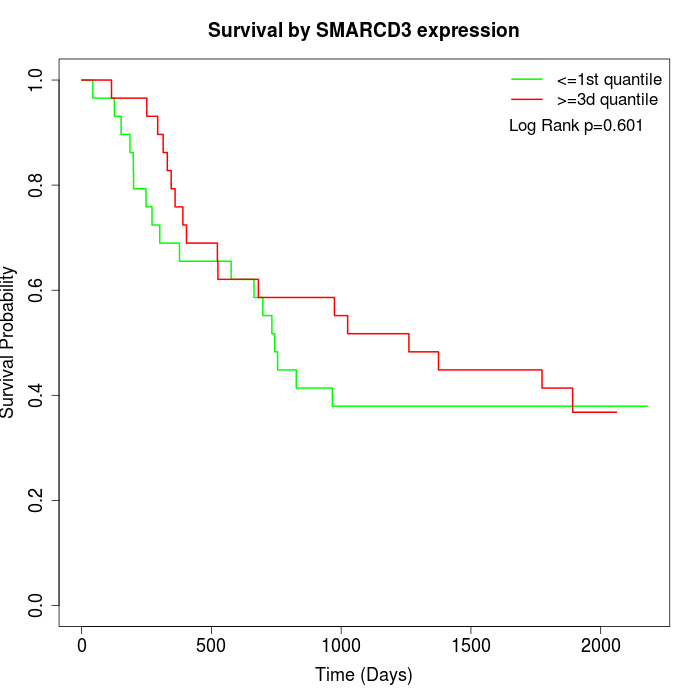
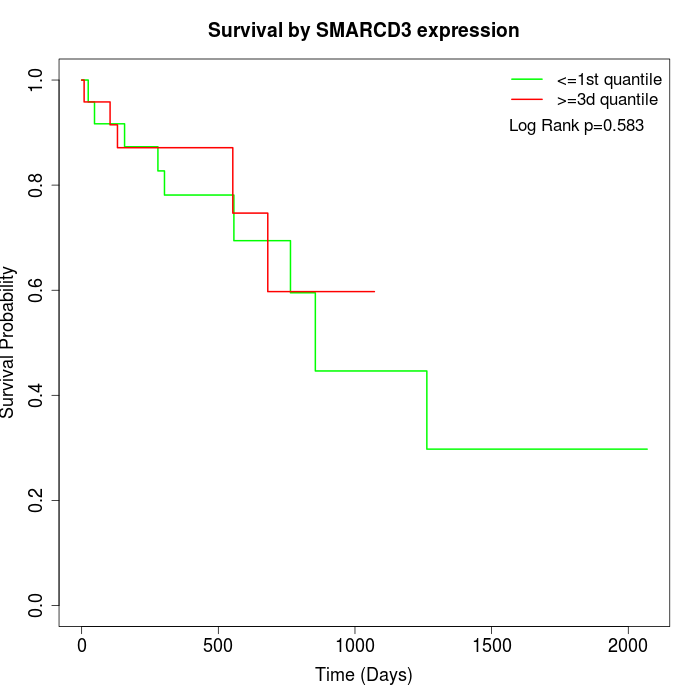
Survival by SMARCD3 expression:
Note: Click image to view full size file.
Copy number change of SMARCD3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SMARCD3 | 6604 | 2 | 4 | 24 | |
| GSE20123 | SMARCD3 | 6604 | 2 | 4 | 24 | |
| GSE43470 | SMARCD3 | 6604 | 2 | 5 | 36 | |
| GSE46452 | SMARCD3 | 6604 | 7 | 2 | 50 | |
| GSE47630 | SMARCD3 | 6604 | 5 | 9 | 26 | |
| GSE54993 | SMARCD3 | 6604 | 3 | 3 | 64 | |
| GSE54994 | SMARCD3 | 6604 | 5 | 8 | 40 | |
| GSE60625 | SMARCD3 | 6604 | 0 | 0 | 11 | |
| GSE74703 | SMARCD3 | 6604 | 2 | 4 | 30 | |
| GSE74704 | SMARCD3 | 6604 | 1 | 4 | 15 | |
| TCGA | SMARCD3 | 6604 | 28 | 25 | 43 |
Total number of gains: 57; Total number of losses: 68; Total Number of normals: 363.
Somatic mutations of SMARCD3:
Generating mutation plots.
Highly correlated genes for SMARCD3:
Showing top 20/638 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SMARCD3 | MOG | 0.757213 | 3 | 0 | 3 |
| SMARCD3 | PSMG3-AS1 | 0.746828 | 3 | 0 | 3 |
| SMARCD3 | RASSF8-AS1 | 0.734414 | 3 | 0 | 3 |
| SMARCD3 | DYSF | 0.717144 | 5 | 0 | 5 |
| SMARCD3 | BVES | 0.700812 | 4 | 0 | 4 |
| SMARCD3 | SLC35F1 | 0.695945 | 3 | 0 | 3 |
| SMARCD3 | TSPAN18 | 0.675013 | 6 | 0 | 4 |
| SMARCD3 | TSPAN7 | 0.672765 | 4 | 0 | 3 |
| SMARCD3 | FOXF1 | 0.669071 | 9 | 0 | 7 |
| SMARCD3 | TMTC1 | 0.668869 | 5 | 0 | 5 |
| SMARCD3 | DIP2C | 0.666799 | 8 | 0 | 7 |
| SMARCD3 | STON1 | 0.666652 | 7 | 0 | 6 |
| SMARCD3 | ZNF423 | 0.665348 | 8 | 0 | 8 |
| SMARCD3 | PAMR1 | 0.665199 | 7 | 0 | 6 |
| SMARCD3 | C1orf198 | 0.661337 | 4 | 0 | 4 |
| SMARCD3 | CNN1 | 0.659171 | 7 | 0 | 6 |
| SMARCD3 | APOD | 0.6588 | 7 | 0 | 7 |
| SMARCD3 | HMCN1 | 0.653971 | 4 | 0 | 3 |
| SMARCD3 | JAM3 | 0.653383 | 9 | 0 | 7 |
| SMARCD3 | MYLK | 0.651226 | 7 | 0 | 6 |
For details and further investigation, click here