| Full name: junctional adhesion molecule 3 | Alias Symbol: JAM-C|JAMC | ||
| Type: protein-coding gene | Cytoband: 11q25 | ||
| Entrez ID: 83700 | HGNC ID: HGNC:15532 | Ensembl Gene: ENSG00000166086 | OMIM ID: 606871 |
| Drug and gene relationship at DGIdb | |||
JAM3 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04530 | Tight junction |
Expression of JAM3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | JAM3 | 83700 | 212813_at | -1.3761 | 0.1706 | |
| GSE20347 | JAM3 | 83700 | 212813_at | -0.1585 | 0.4035 | |
| GSE23400 | JAM3 | 83700 | 212813_at | -0.4915 | 0.0011 | |
| GSE26886 | JAM3 | 83700 | 212813_at | 1.3265 | 0.0032 | |
| GSE29001 | JAM3 | 83700 | 212813_at | -0.1082 | 0.7843 | |
| GSE38129 | JAM3 | 83700 | 212813_at | -0.7181 | 0.0863 | |
| GSE45670 | JAM3 | 83700 | 212813_at | -2.0254 | 0.0000 | |
| GSE53622 | JAM3 | 83700 | 25483 | -0.6481 | 0.0001 | |
| GSE53624 | JAM3 | 83700 | 25483 | -0.5716 | 0.0000 | |
| GSE63941 | JAM3 | 83700 | 212813_at | -4.2208 | 0.0008 | |
| GSE77861 | JAM3 | 83700 | 212813_at | 0.1889 | 0.3425 | |
| GSE97050 | JAM3 | 83700 | A_24_P86993 | -0.8752 | 0.1626 | |
| SRP007169 | JAM3 | 83700 | RNAseq | 3.8507 | 0.0021 | |
| SRP064894 | JAM3 | 83700 | RNAseq | -0.1631 | 0.6808 | |
| SRP133303 | JAM3 | 83700 | RNAseq | -0.2614 | 0.5398 | |
| SRP159526 | JAM3 | 83700 | RNAseq | -0.1361 | 0.7766 | |
| SRP193095 | JAM3 | 83700 | RNAseq | 0.5187 | 0.0697 | |
| SRP219564 | JAM3 | 83700 | RNAseq | -0.9449 | 0.3919 | |
| TCGA | JAM3 | 83700 | RNAseq | -0.5476 | 0.0000 |
Upregulated datasets: 2; Downregulated datasets: 2.
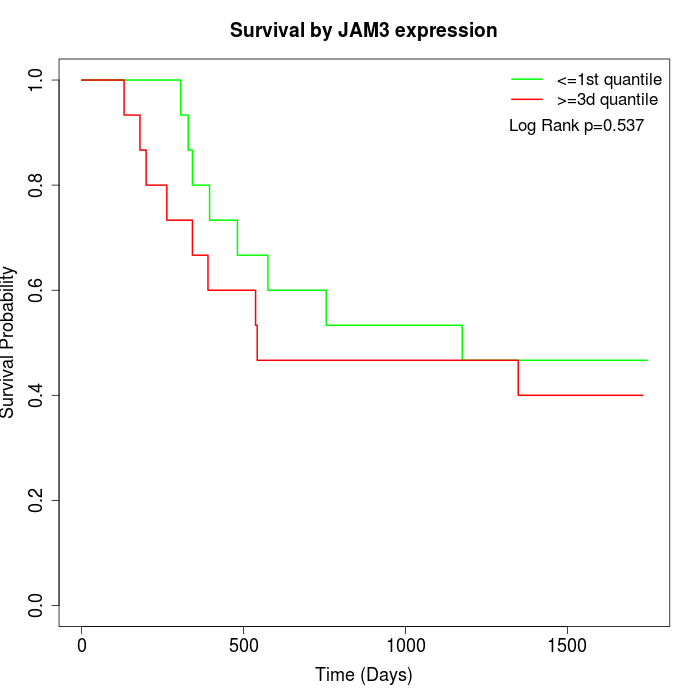
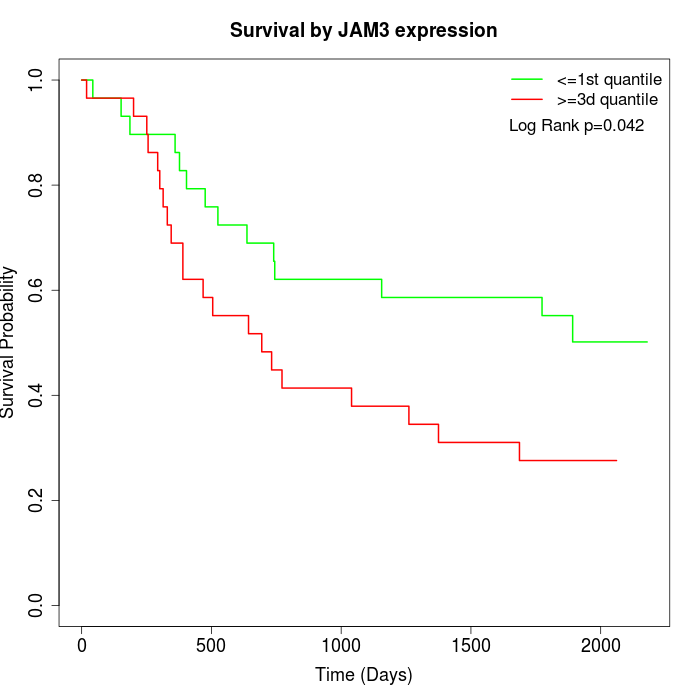
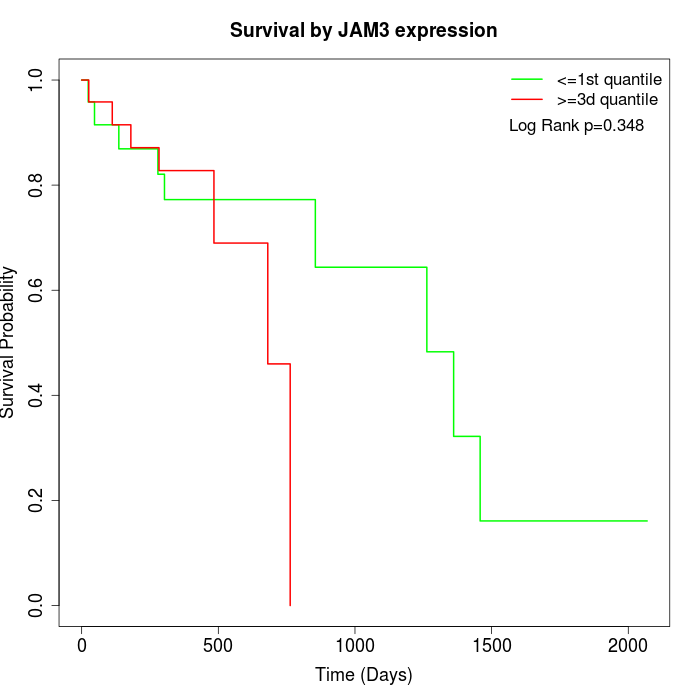
Survival by JAM3 expression:
Note: Click image to view full size file.
Copy number change of JAM3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | JAM3 | 83700 | 0 | 15 | 15 | |
| GSE20123 | JAM3 | 83700 | 0 | 15 | 15 | |
| GSE43470 | JAM3 | 83700 | 2 | 8 | 33 | |
| GSE46452 | JAM3 | 83700 | 3 | 26 | 30 | |
| GSE47630 | JAM3 | 83700 | 2 | 19 | 19 | |
| GSE54993 | JAM3 | 83700 | 10 | 0 | 60 | |
| GSE54994 | JAM3 | 83700 | 5 | 18 | 30 | |
| GSE60625 | JAM3 | 83700 | 0 | 3 | 8 | |
| GSE74703 | JAM3 | 83700 | 1 | 5 | 30 | |
| GSE74704 | JAM3 | 83700 | 0 | 11 | 9 | |
| TCGA | JAM3 | 83700 | 6 | 49 | 41 |
Total number of gains: 29; Total number of losses: 169; Total Number of normals: 290.
Somatic mutations of JAM3:
Generating mutation plots.
Highly correlated genes for JAM3:
Showing top 20/1418 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| JAM3 | MSRB3 | 0.878464 | 7 | 0 | 7 |
| JAM3 | FIBIN | 0.83777 | 5 | 0 | 5 |
| JAM3 | MAGI2-AS3 | 0.828011 | 6 | 0 | 6 |
| JAM3 | KANK2 | 0.823295 | 11 | 0 | 10 |
| JAM3 | TBX5 | 0.82312 | 7 | 0 | 7 |
| JAM3 | FOXF1 | 0.821175 | 13 | 0 | 13 |
| JAM3 | RUNX1T1 | 0.820418 | 8 | 0 | 8 |
| JAM3 | C11orf96 | 0.820188 | 5 | 0 | 5 |
| JAM3 | PREX2 | 0.81825 | 4 | 0 | 4 |
| JAM3 | TMEM47 | 0.816968 | 13 | 0 | 13 |
| JAM3 | NFASC | 0.81661 | 6 | 0 | 6 |
| JAM3 | TCEAL5 | 0.814094 | 3 | 0 | 3 |
| JAM3 | DACT3 | 0.813521 | 7 | 0 | 7 |
| JAM3 | ZEB1 | 0.808387 | 3 | 0 | 3 |
| JAM3 | FERMT2 | 0.804294 | 13 | 0 | 13 |
| JAM3 | DDR2 | 0.803294 | 13 | 0 | 13 |
| JAM3 | TCEAL6 | 0.8032 | 3 | 0 | 3 |
| JAM3 | PDE5A | 0.800929 | 10 | 0 | 10 |
| JAM3 | PDLIM3 | 0.797503 | 13 | 0 | 13 |
| JAM3 | RASSF3 | 0.797058 | 4 | 0 | 4 |
For details and further investigation, click here