| Full name: superoxide dismutase 1 | Alias Symbol: IPOA | ||
| Type: protein-coding gene | Cytoband: 21q22.11 | ||
| Entrez ID: 6647 | HGNC ID: HGNC:11179 | Ensembl Gene: ENSG00000142168 | OMIM ID: 147450 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
SOD1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04213 | Longevity regulating pathway - multiple species | |
| hsa05014 | Amyotrophic lateral sclerosis (ALS) | |
| hsa05020 | Prion diseases |
Expression of SOD1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SOD1 | 6647 | 200642_at | 0.0425 | 0.9176 | |
| GSE20347 | SOD1 | 6647 | 200642_at | 0.1724 | 0.1930 | |
| GSE23400 | SOD1 | 6647 | 200642_at | 0.2032 | 0.0189 | |
| GSE26886 | SOD1 | 6647 | 200642_at | -0.4275 | 0.0779 | |
| GSE29001 | SOD1 | 6647 | 200642_at | 0.0593 | 0.7781 | |
| GSE38129 | SOD1 | 6647 | 200642_at | 0.1935 | 0.1058 | |
| GSE45670 | SOD1 | 6647 | 200642_at | -0.0962 | 0.5709 | |
| GSE53622 | SOD1 | 6647 | 66447 | -0.1811 | 0.0023 | |
| GSE53624 | SOD1 | 6647 | 66447 | 0.3023 | 0.0000 | |
| GSE63941 | SOD1 | 6647 | 200642_at | -0.2076 | 0.6221 | |
| GSE77861 | SOD1 | 6647 | 200642_at | -0.0842 | 0.7094 | |
| GSE97050 | SOD1 | 6647 | A_23_P154840 | -0.5885 | 0.0684 | |
| SRP007169 | SOD1 | 6647 | RNAseq | -1.0133 | 0.0120 | |
| SRP008496 | SOD1 | 6647 | RNAseq | -0.3908 | 0.2213 | |
| SRP064894 | SOD1 | 6647 | RNAseq | 0.2281 | 0.3642 | |
| SRP133303 | SOD1 | 6647 | RNAseq | -0.1984 | 0.3029 | |
| SRP159526 | SOD1 | 6647 | RNAseq | 0.5697 | 0.0211 | |
| SRP193095 | SOD1 | 6647 | RNAseq | -0.5196 | 0.0010 | |
| SRP219564 | SOD1 | 6647 | RNAseq | -0.4081 | 0.1775 | |
| TCGA | SOD1 | 6647 | RNAseq | 0.0429 | 0.4132 |
Upregulated datasets: 0; Downregulated datasets: 1.
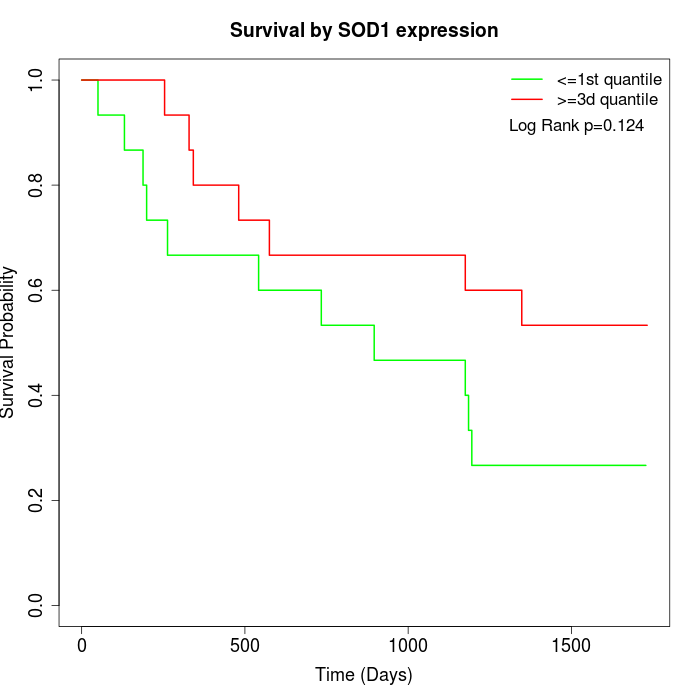
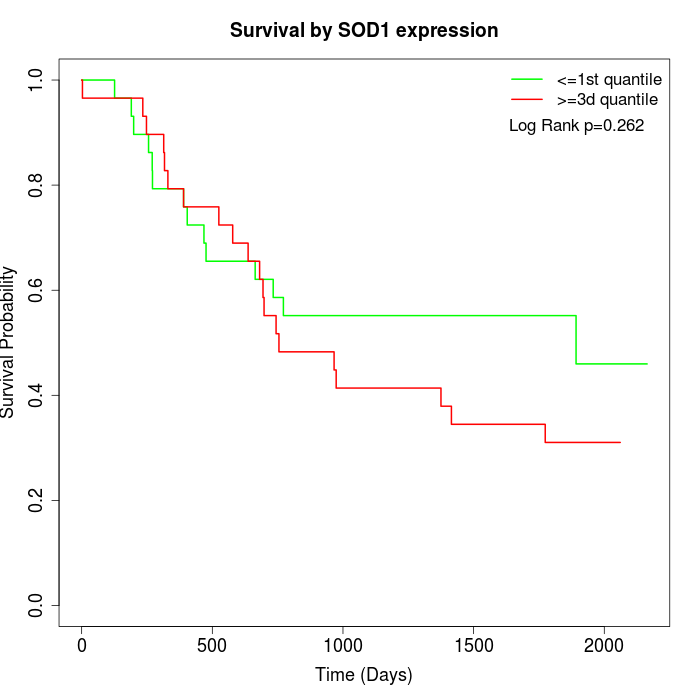
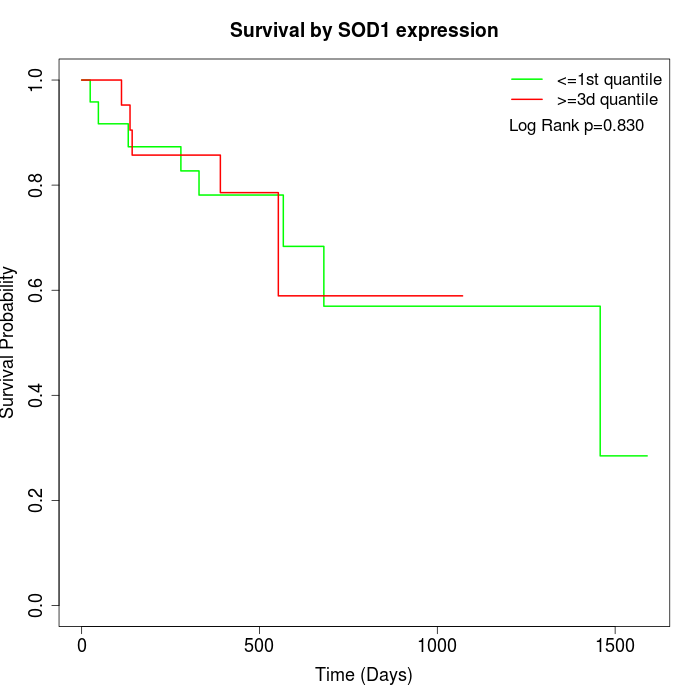
Survival by SOD1 expression:
Note: Click image to view full size file.
Copy number change of SOD1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SOD1 | 6647 | 3 | 10 | 17 | |
| GSE20123 | SOD1 | 6647 | 3 | 10 | 17 | |
| GSE43470 | SOD1 | 6647 | 2 | 9 | 32 | |
| GSE46452 | SOD1 | 6647 | 1 | 21 | 37 | |
| GSE47630 | SOD1 | 6647 | 6 | 17 | 17 | |
| GSE54993 | SOD1 | 6647 | 9 | 1 | 60 | |
| GSE54994 | SOD1 | 6647 | 1 | 9 | 43 | |
| GSE60625 | SOD1 | 6647 | 0 | 0 | 11 | |
| GSE74703 | SOD1 | 6647 | 2 | 6 | 28 | |
| GSE74704 | SOD1 | 6647 | 2 | 6 | 12 | |
| TCGA | SOD1 | 6647 | 8 | 37 | 51 |
Total number of gains: 37; Total number of losses: 126; Total Number of normals: 325.
Somatic mutations of SOD1:
Generating mutation plots.
Highly correlated genes for SOD1:
Showing top 20/134 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SOD1 | CNST | 0.693983 | 3 | 0 | 3 |
| SOD1 | TBRG1 | 0.693487 | 3 | 0 | 3 |
| SOD1 | MLLT6 | 0.68164 | 4 | 0 | 4 |
| SOD1 | RCOR3 | 0.679728 | 3 | 0 | 3 |
| SOD1 | FAM193A | 0.679721 | 4 | 0 | 3 |
| SOD1 | GABPB2 | 0.654476 | 3 | 0 | 3 |
| SOD1 | ZNF784 | 0.650174 | 3 | 0 | 3 |
| SOD1 | RCBTB1 | 0.630561 | 3 | 0 | 3 |
| SOD1 | NUB1 | 0.623614 | 3 | 0 | 3 |
| SOD1 | CTSH | 0.622146 | 3 | 0 | 3 |
| SOD1 | ZNF420 | 0.617345 | 3 | 0 | 3 |
| SOD1 | MDN1 | 0.613 | 3 | 0 | 3 |
| SOD1 | ARL1 | 0.610661 | 4 | 0 | 3 |
| SOD1 | MOCS2 | 0.609752 | 4 | 0 | 3 |
| SOD1 | RASSF4 | 0.608321 | 3 | 0 | 3 |
| SOD1 | RBBP4 | 0.60825 | 6 | 0 | 4 |
| SOD1 | CTDSPL | 0.606693 | 5 | 0 | 3 |
| SOD1 | CREG1 | 0.605973 | 7 | 0 | 4 |
| SOD1 | DNTTIP2 | 0.605172 | 5 | 0 | 3 |
| SOD1 | DZIP3 | 0.602536 | 4 | 0 | 3 |
For details and further investigation, click here