| Full name: cathepsin H | Alias Symbol: ACC-4|ACC-5|ACC4|ACC5 | ||
| Type: protein-coding gene | Cytoband: 15q25.1 | ||
| Entrez ID: 1512 | HGNC ID: HGNC:2535 | Ensembl Gene: ENSG00000103811 | OMIM ID: 116820 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
CTSH involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04210 | Apoptosis |
Expression of CTSH:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CTSH | 1512 | 202295_s_at | 0.4259 | 0.5163 | |
| GSE20347 | CTSH | 1512 | 202295_s_at | 0.0907 | 0.7622 | |
| GSE23400 | CTSH | 1512 | 202295_s_at | 0.2971 | 0.0080 | |
| GSE26886 | CTSH | 1512 | 202295_s_at | 0.4187 | 0.3632 | |
| GSE29001 | CTSH | 1512 | 202295_s_at | 0.4050 | 0.3011 | |
| GSE38129 | CTSH | 1512 | 202295_s_at | 0.3482 | 0.1782 | |
| GSE45670 | CTSH | 1512 | 202295_s_at | 0.1914 | 0.5676 | |
| GSE53622 | CTSH | 1512 | 138084 | 0.3711 | 0.0021 | |
| GSE53624 | CTSH | 1512 | 138084 | 0.2056 | 0.0460 | |
| GSE63941 | CTSH | 1512 | 202295_s_at | 2.1137 | 0.2249 | |
| GSE77861 | CTSH | 1512 | 202295_s_at | -0.2086 | 0.5817 | |
| GSE97050 | CTSH | 1512 | A_23_P14774 | 0.8071 | 0.1049 | |
| SRP007169 | CTSH | 1512 | RNAseq | -0.4348 | 0.4077 | |
| SRP008496 | CTSH | 1512 | RNAseq | -0.2440 | 0.4304 | |
| SRP064894 | CTSH | 1512 | RNAseq | 0.8612 | 0.0029 | |
| SRP133303 | CTSH | 1512 | RNAseq | 0.7793 | 0.0001 | |
| SRP159526 | CTSH | 1512 | RNAseq | 0.3078 | 0.5721 | |
| SRP193095 | CTSH | 1512 | RNAseq | 0.0914 | 0.7159 | |
| SRP219564 | CTSH | 1512 | RNAseq | 1.1196 | 0.0017 | |
| TCGA | CTSH | 1512 | RNAseq | -0.1622 | 0.0774 |
Upregulated datasets: 1; Downregulated datasets: 0.
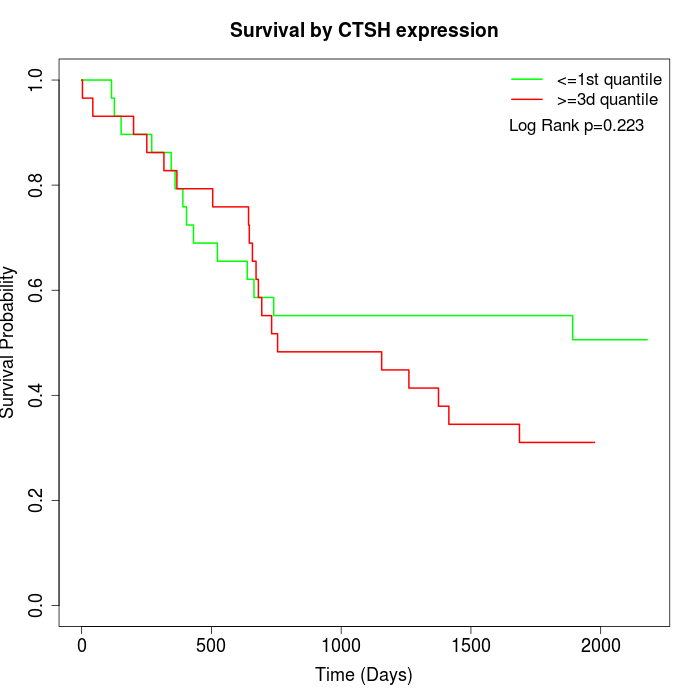
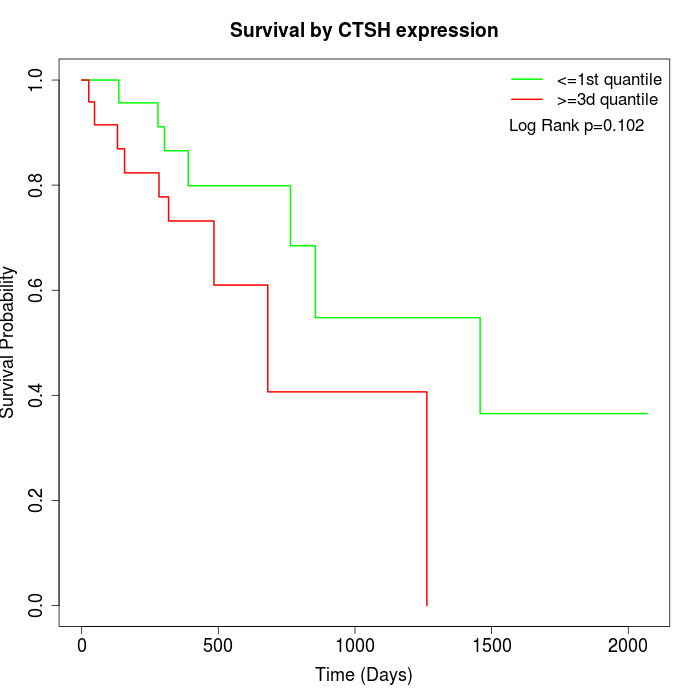
Survival by CTSH expression:
Note: Click image to view full size file.
Copy number change of CTSH:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CTSH | 1512 | 7 | 1 | 22 | |
| GSE20123 | CTSH | 1512 | 7 | 1 | 22 | |
| GSE43470 | CTSH | 1512 | 4 | 5 | 34 | |
| GSE46452 | CTSH | 1512 | 3 | 7 | 49 | |
| GSE47630 | CTSH | 1512 | 8 | 10 | 22 | |
| GSE54993 | CTSH | 1512 | 5 | 6 | 59 | |
| GSE54994 | CTSH | 1512 | 5 | 6 | 42 | |
| GSE60625 | CTSH | 1512 | 4 | 0 | 7 | |
| GSE74703 | CTSH | 1512 | 4 | 3 | 29 | |
| GSE74704 | CTSH | 1512 | 3 | 1 | 16 | |
| TCGA | CTSH | 1512 | 14 | 15 | 67 |
Total number of gains: 64; Total number of losses: 55; Total Number of normals: 369.
Somatic mutations of CTSH:
Generating mutation plots.
Highly correlated genes for CTSH:
Showing top 20/250 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CTSH | SLC28A3 | 0.783203 | 3 | 0 | 3 |
| CTSH | PREX1 | 0.779135 | 3 | 0 | 3 |
| CTSH | TCP11L1 | 0.741239 | 3 | 0 | 3 |
| CTSH | AKIP1 | 0.734159 | 3 | 0 | 3 |
| CTSH | FAM49A | 0.717462 | 4 | 0 | 4 |
| CTSH | TBC1D2 | 0.711943 | 3 | 0 | 3 |
| CTSH | TGFB1 | 0.704935 | 3 | 0 | 3 |
| CTSH | CERKL | 0.702653 | 3 | 0 | 3 |
| CTSH | FLOT2 | 0.702053 | 3 | 0 | 3 |
| CTSH | CDT1 | 0.699491 | 3 | 0 | 3 |
| CTSH | GLI3 | 0.69905 | 3 | 0 | 3 |
| CTSH | A1BG | 0.689238 | 3 | 0 | 3 |
| CTSH | FYN | 0.687544 | 4 | 0 | 3 |
| CTSH | AK2 | 0.681938 | 3 | 0 | 3 |
| CTSH | WDR86 | 0.681902 | 3 | 0 | 3 |
| CTSH | CBX1 | 0.681201 | 3 | 0 | 3 |
| CTSH | VNN2 | 0.67641 | 4 | 0 | 3 |
| CTSH | RASAL3 | 0.674356 | 3 | 0 | 3 |
| CTSH | FKBP10 | 0.670625 | 3 | 0 | 3 |
| CTSH | NDN | 0.6589 | 3 | 0 | 3 |
For details and further investigation, click here