| Full name: transcription factor 7 like 2 | Alias Symbol: TCF-4 | ||
| Type: protein-coding gene | Cytoband: 10q25.2-q25.3 | ||
| Entrez ID: 6934 | HGNC ID: HGNC:11641 | Ensembl Gene: ENSG00000148737 | OMIM ID: 602228 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
TCF7L2 involved pathways:
Expression of TCF7L2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TCF7L2 | 6934 | 212761_at | -0.2271 | 0.6137 | |
| GSE20347 | TCF7L2 | 6934 | 212761_at | 0.0423 | 0.8700 | |
| GSE23400 | TCF7L2 | 6934 | 212761_at | 0.0691 | 0.4936 | |
| GSE26886 | TCF7L2 | 6934 | 212761_at | 0.9331 | 0.0003 | |
| GSE29001 | TCF7L2 | 6934 | 212761_at | 0.2155 | 0.2700 | |
| GSE38129 | TCF7L2 | 6934 | 212761_at | -0.1382 | 0.4370 | |
| GSE45670 | TCF7L2 | 6934 | 212761_at | -0.7024 | 0.0003 | |
| GSE53622 | TCF7L2 | 6934 | 48686 | -0.0161 | 0.8622 | |
| GSE53624 | TCF7L2 | 6934 | 120084 | 0.1214 | 0.0640 | |
| GSE63941 | TCF7L2 | 6934 | 212761_at | -0.4185 | 0.3194 | |
| GSE77861 | TCF7L2 | 6934 | 212761_at | 0.5021 | 0.2390 | |
| GSE97050 | TCF7L2 | 6934 | A_33_P3377304 | 0.2864 | 0.2942 | |
| SRP007169 | TCF7L2 | 6934 | RNAseq | 0.6789 | 0.1502 | |
| SRP008496 | TCF7L2 | 6934 | RNAseq | 0.9984 | 0.0047 | |
| SRP064894 | TCF7L2 | 6934 | RNAseq | -0.1080 | 0.5118 | |
| SRP133303 | TCF7L2 | 6934 | RNAseq | 0.1446 | 0.4007 | |
| SRP159526 | TCF7L2 | 6934 | RNAseq | 0.0328 | 0.8905 | |
| SRP193095 | TCF7L2 | 6934 | RNAseq | 0.1933 | 0.4885 | |
| SRP219564 | TCF7L2 | 6934 | RNAseq | 0.1130 | 0.6726 | |
| TCGA | TCF7L2 | 6934 | RNAseq | -0.1306 | 0.0260 |
Upregulated datasets: 0; Downregulated datasets: 0.
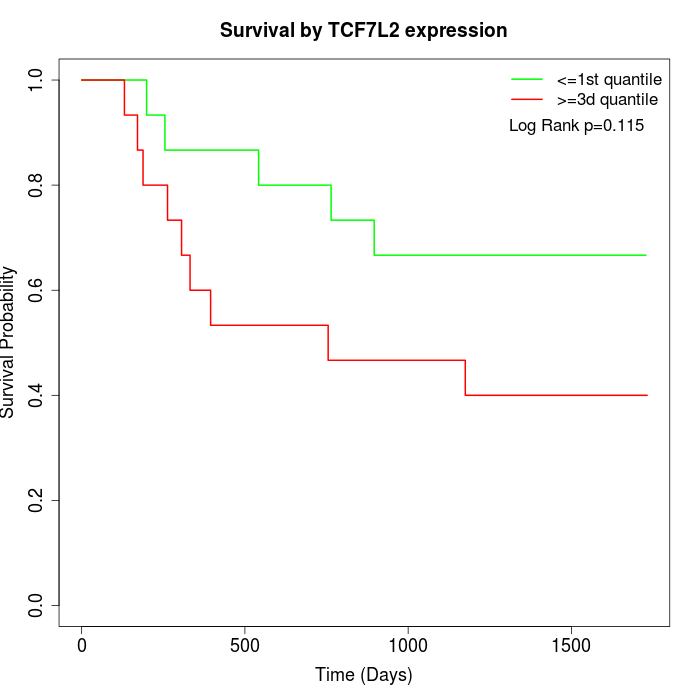
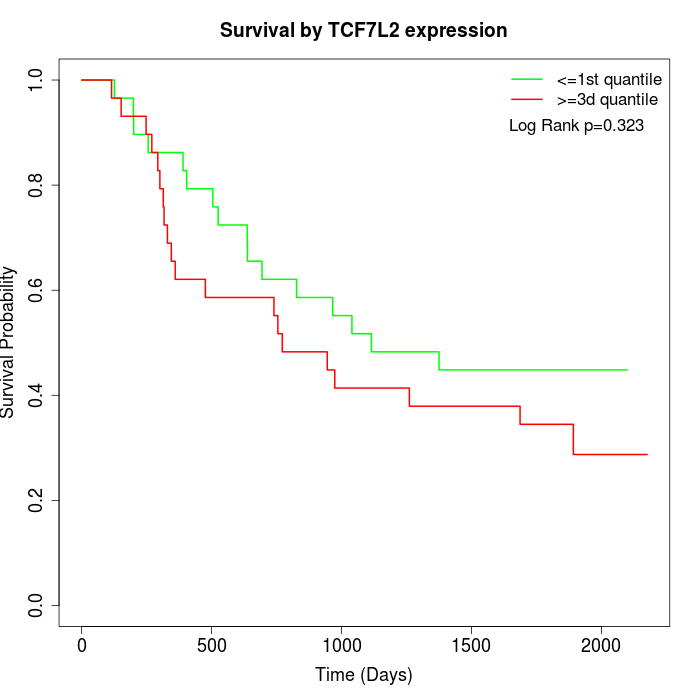
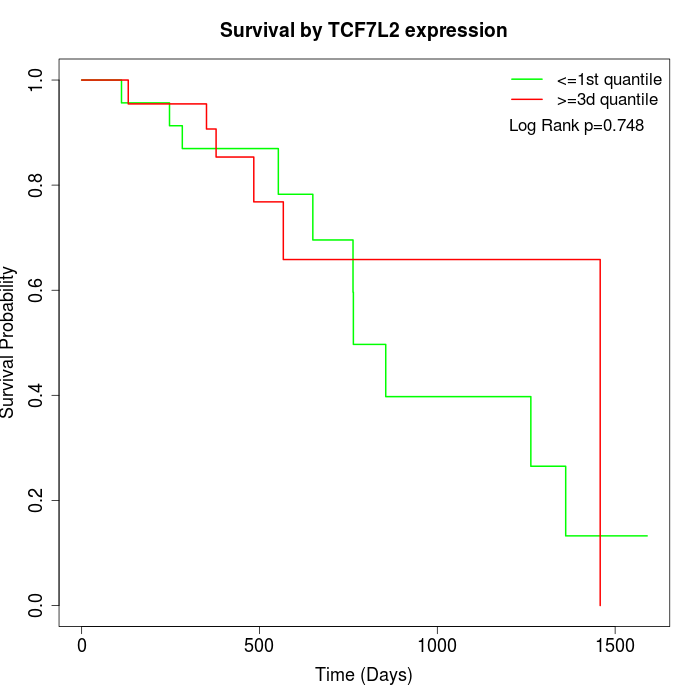
Survival by TCF7L2 expression:
Note: Click image to view full size file.
Copy number change of TCF7L2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TCF7L2 | 6934 | 1 | 8 | 21 | |
| GSE20123 | TCF7L2 | 6934 | 1 | 7 | 22 | |
| GSE43470 | TCF7L2 | 6934 | 0 | 9 | 34 | |
| GSE46452 | TCF7L2 | 6934 | 0 | 11 | 48 | |
| GSE47630 | TCF7L2 | 6934 | 2 | 14 | 24 | |
| GSE54993 | TCF7L2 | 6934 | 8 | 0 | 62 | |
| GSE54994 | TCF7L2 | 6934 | 1 | 10 | 42 | |
| GSE60625 | TCF7L2 | 6934 | 0 | 0 | 11 | |
| GSE74703 | TCF7L2 | 6934 | 0 | 6 | 30 | |
| GSE74704 | TCF7L2 | 6934 | 1 | 3 | 16 | |
| TCGA | TCF7L2 | 6934 | 4 | 29 | 63 |
Total number of gains: 18; Total number of losses: 97; Total Number of normals: 373.
Somatic mutations of TCF7L2:
Generating mutation plots.
Highly correlated genes for TCF7L2:
Showing top 20/496 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TCF7L2 | TMED8 | 0.808124 | 3 | 0 | 3 |
| TCF7L2 | EIF2D | 0.805361 | 3 | 0 | 3 |
| TCF7L2 | HNRNPA1 | 0.798493 | 3 | 0 | 3 |
| TCF7L2 | ANAPC10 | 0.792701 | 3 | 0 | 3 |
| TCF7L2 | SPICE1 | 0.786724 | 3 | 0 | 3 |
| TCF7L2 | SLC39A10 | 0.77263 | 3 | 0 | 3 |
| TCF7L2 | ZFP90 | 0.768234 | 3 | 0 | 3 |
| TCF7L2 | ZNF44 | 0.762286 | 3 | 0 | 3 |
| TCF7L2 | ZNF766 | 0.759571 | 3 | 0 | 3 |
| TCF7L2 | BRD9 | 0.759505 | 3 | 0 | 3 |
| TCF7L2 | POLR2J | 0.757108 | 3 | 0 | 3 |
| TCF7L2 | ZCCHC7 | 0.756017 | 3 | 0 | 3 |
| TCF7L2 | EFCAB7 | 0.753628 | 4 | 0 | 4 |
| TCF7L2 | TATDN1 | 0.750083 | 3 | 0 | 3 |
| TCF7L2 | TIMM9 | 0.749918 | 3 | 0 | 3 |
| TCF7L2 | NSMCE2 | 0.736523 | 3 | 0 | 3 |
| TCF7L2 | DOCK5 | 0.736173 | 3 | 0 | 3 |
| TCF7L2 | TIMP1 | 0.736023 | 3 | 0 | 3 |
| TCF7L2 | ZNF615 | 0.733667 | 3 | 0 | 3 |
| TCF7L2 | SIX4 | 0.732206 | 3 | 0 | 3 |
For details and further investigation, click here