| Full name: anaphase promoting complex subunit 10 | Alias Symbol: APC10|DOC1|DKFZP564L0562 | ||
| Type: protein-coding gene | Cytoband: 4q31.21 | ||
| Entrez ID: 10393 | HGNC ID: HGNC:24077 | Ensembl Gene: ENSG00000164162 | OMIM ID: 613745 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ANAPC10 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04110 | Cell cycle | |
| hsa04114 | Oocyte meiosis | |
| hsa04914 | Progesterone-mediated oocyte maturation | |
| hsa05166 | HTLV-I infection |
Expression of ANAPC10:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ANAPC10 | 10393 | 207845_s_at | 0.3239 | 0.3083 | |
| GSE20347 | ANAPC10 | 10393 | 207845_s_at | 0.3250 | 0.1383 | |
| GSE23400 | ANAPC10 | 10393 | 207845_s_at | 0.2812 | 0.0000 | |
| GSE26886 | ANAPC10 | 10393 | 207845_s_at | 1.7539 | 0.0000 | |
| GSE29001 | ANAPC10 | 10393 | 207845_s_at | 0.4097 | 0.4091 | |
| GSE38129 | ANAPC10 | 10393 | 207845_s_at | 0.3720 | 0.0292 | |
| GSE45670 | ANAPC10 | 10393 | 207845_s_at | 0.1210 | 0.5330 | |
| GSE53622 | ANAPC10 | 10393 | 52795 | 0.4029 | 0.0000 | |
| GSE53624 | ANAPC10 | 10393 | 52795 | 0.5042 | 0.0000 | |
| GSE63941 | ANAPC10 | 10393 | 207845_s_at | 0.2379 | 0.4964 | |
| GSE77861 | ANAPC10 | 10393 | 207845_s_at | 0.2103 | 0.5032 | |
| GSE97050 | ANAPC10 | 10393 | A_23_P250994 | 0.4881 | 0.2461 | |
| SRP007169 | ANAPC10 | 10393 | RNAseq | 0.0515 | 0.9096 | |
| SRP008496 | ANAPC10 | 10393 | RNAseq | 0.1411 | 0.6256 | |
| SRP064894 | ANAPC10 | 10393 | RNAseq | 0.2357 | 0.1459 | |
| SRP133303 | ANAPC10 | 10393 | RNAseq | 0.7687 | 0.0001 | |
| SRP159526 | ANAPC10 | 10393 | RNAseq | 0.3479 | 0.1970 | |
| SRP193095 | ANAPC10 | 10393 | RNAseq | 0.2971 | 0.0194 | |
| SRP219564 | ANAPC10 | 10393 | RNAseq | 0.0725 | 0.8344 | |
| TCGA | ANAPC10 | 10393 | RNAseq | 0.2660 | 0.0006 |
Upregulated datasets: 1; Downregulated datasets: 0.
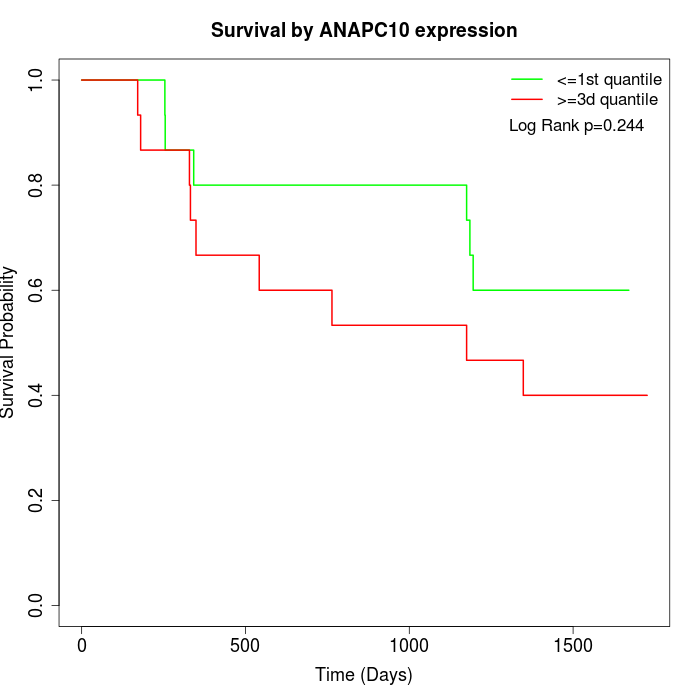
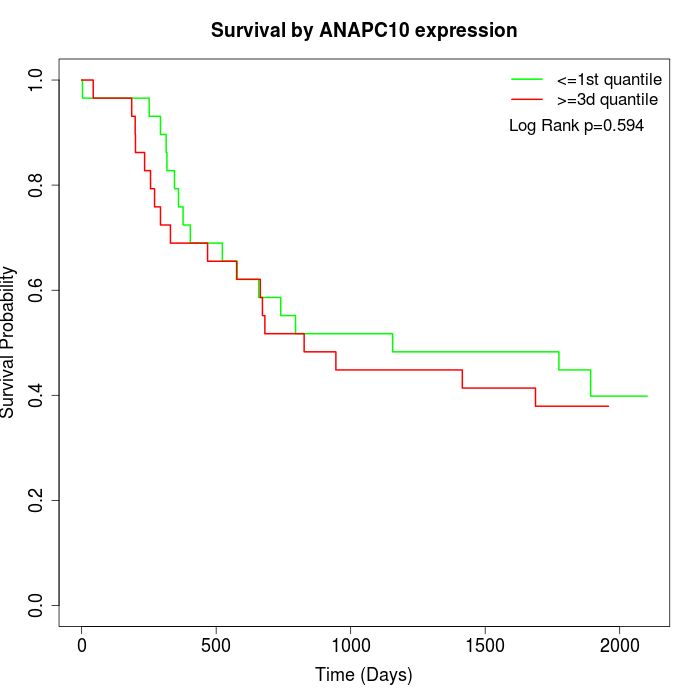
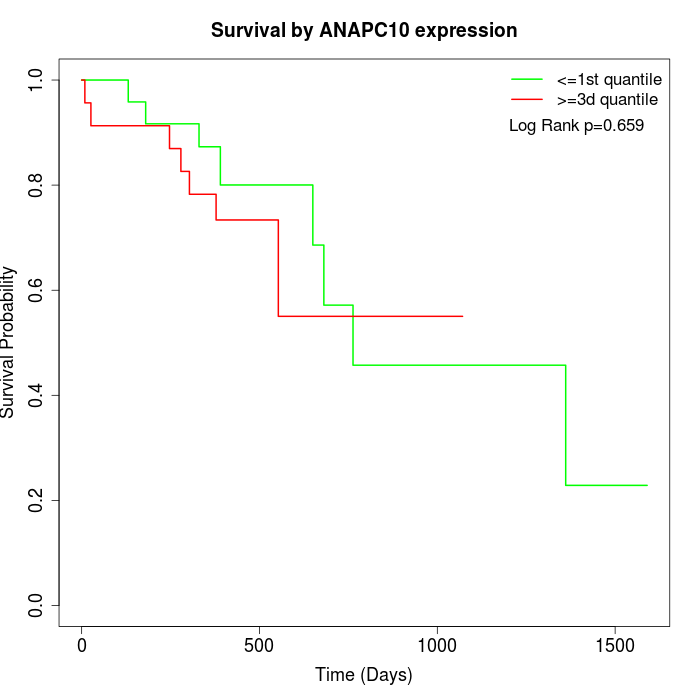
Survival by ANAPC10 expression:
Note: Click image to view full size file.
Copy number change of ANAPC10:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ANAPC10 | 10393 | 0 | 13 | 17 | |
| GSE20123 | ANAPC10 | 10393 | 0 | 13 | 17 | |
| GSE43470 | ANAPC10 | 10393 | 0 | 13 | 30 | |
| GSE46452 | ANAPC10 | 10393 | 1 | 36 | 22 | |
| GSE47630 | ANAPC10 | 10393 | 0 | 22 | 18 | |
| GSE54993 | ANAPC10 | 10393 | 10 | 0 | 60 | |
| GSE54994 | ANAPC10 | 10393 | 2 | 11 | 40 | |
| GSE60625 | ANAPC10 | 10393 | 0 | 1 | 10 | |
| GSE74703 | ANAPC10 | 10393 | 0 | 11 | 25 | |
| GSE74704 | ANAPC10 | 10393 | 0 | 7 | 13 | |
| TCGA | ANAPC10 | 10393 | 11 | 32 | 53 |
Total number of gains: 24; Total number of losses: 159; Total Number of normals: 305.
Somatic mutations of ANAPC10:
Generating mutation plots.
Highly correlated genes for ANAPC10:
Showing top 20/1444 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ANAPC10 | PET117 | 0.908913 | 3 | 0 | 3 |
| ANAPC10 | TCF7L2 | 0.792701 | 3 | 0 | 3 |
| ANAPC10 | ROMO1 | 0.789807 | 3 | 0 | 3 |
| ANAPC10 | MSTO1 | 0.781312 | 3 | 0 | 3 |
| ANAPC10 | CMC1 | 0.780443 | 3 | 0 | 3 |
| ANAPC10 | CENPBD1 | 0.779718 | 3 | 0 | 3 |
| ANAPC10 | ZNF627 | 0.775717 | 3 | 0 | 3 |
| ANAPC10 | EIF3L | 0.762082 | 3 | 0 | 3 |
| ANAPC10 | SGMS1 | 0.756508 | 3 | 0 | 3 |
| ANAPC10 | CD44 | 0.742232 | 4 | 0 | 4 |
| ANAPC10 | CCNT1 | 0.730228 | 3 | 0 | 3 |
| ANAPC10 | ARFGEF2 | 0.729341 | 4 | 0 | 3 |
| ANAPC10 | ABCE1 | 0.722964 | 12 | 0 | 12 |
| ANAPC10 | NADK | 0.717972 | 4 | 0 | 4 |
| ANAPC10 | POM121 | 0.717466 | 3 | 0 | 3 |
| ANAPC10 | DNTTIP1 | 0.714654 | 6 | 0 | 5 |
| ANAPC10 | LRRC58 | 0.712335 | 4 | 0 | 3 |
| ANAPC10 | WDR54 | 0.712097 | 4 | 0 | 3 |
| ANAPC10 | LY75 | 0.712067 | 4 | 0 | 4 |
| ANAPC10 | FAM161A | 0.710613 | 4 | 0 | 3 |
For details and further investigation, click here