| Full name: transferrin | Alias Symbol: PRO1557|PRO2086 | ||
| Type: protein-coding gene | Cytoband: 3q22.1 | ||
| Entrez ID: 7018 | HGNC ID: HGNC:11740 | Ensembl Gene: ENSG00000091513 | OMIM ID: 190000 |
| Drug and gene relationship at DGIdb | |||
TF involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04066 | HIF-1 signaling pathway |
Expression of TF:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TF | 7018 | 214063_s_at | -1.6933 | 0.3249 | |
| GSE20347 | TF | 7018 | 214063_s_at | -1.4112 | 0.0216 | |
| GSE23400 | TF | 7018 | 214063_s_at | -0.9701 | 0.0000 | |
| GSE26886 | TF | 7018 | 214063_s_at | -3.8167 | 0.0000 | |
| GSE29001 | TF | 7018 | 214063_s_at | -1.5764 | 0.1201 | |
| GSE38129 | TF | 7018 | 214063_s_at | -1.3298 | 0.0013 | |
| GSE45670 | TF | 7018 | 214063_s_at | -1.4975 | 0.0539 | |
| GSE53622 | TF | 7018 | 19469 | -1.3737 | 0.0000 | |
| GSE53624 | TF | 7018 | 19469 | -2.7449 | 0.0000 | |
| GSE63941 | TF | 7018 | 214064_at | 0.3026 | 0.0132 | |
| GSE77861 | TF | 7018 | 214063_s_at | -2.8486 | 0.0033 | |
| GSE97050 | TF | 7018 | A_23_P212508 | -0.1215 | 0.7440 | |
| SRP007169 | TF | 7018 | RNAseq | 0.3272 | 0.6367 | |
| SRP008496 | TF | 7018 | RNAseq | -0.3916 | 0.4513 | |
| SRP064894 | TF | 7018 | RNAseq | -0.0292 | 0.9360 | |
| SRP133303 | TF | 7018 | RNAseq | 0.0511 | 0.6642 | |
| SRP159526 | TF | 7018 | RNAseq | 1.6294 | 0.0000 | |
| SRP193095 | TF | 7018 | RNAseq | 0.3157 | 0.0629 | |
| SRP219564 | TF | 7018 | RNAseq | 0.2078 | 0.6478 | |
| TCGA | TF | 7018 | RNAseq | -1.2782 | 0.0307 |
Upregulated datasets: 1; Downregulated datasets: 7.
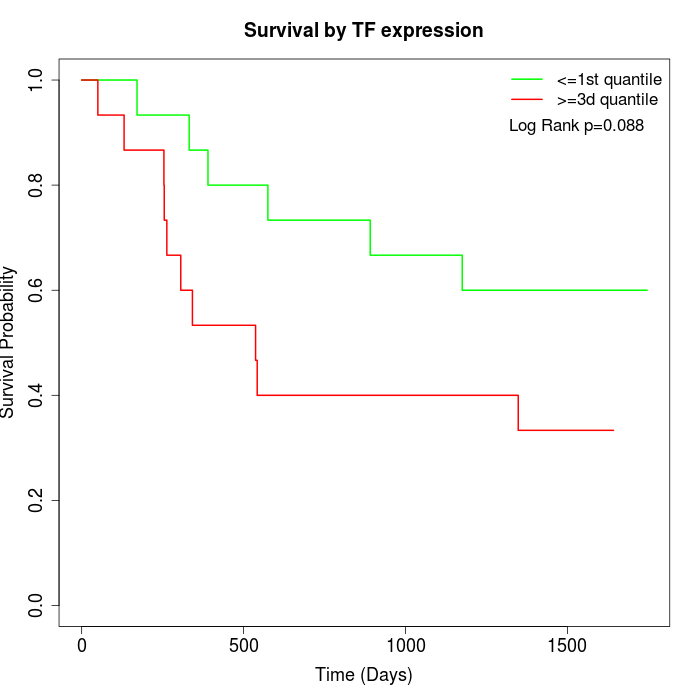
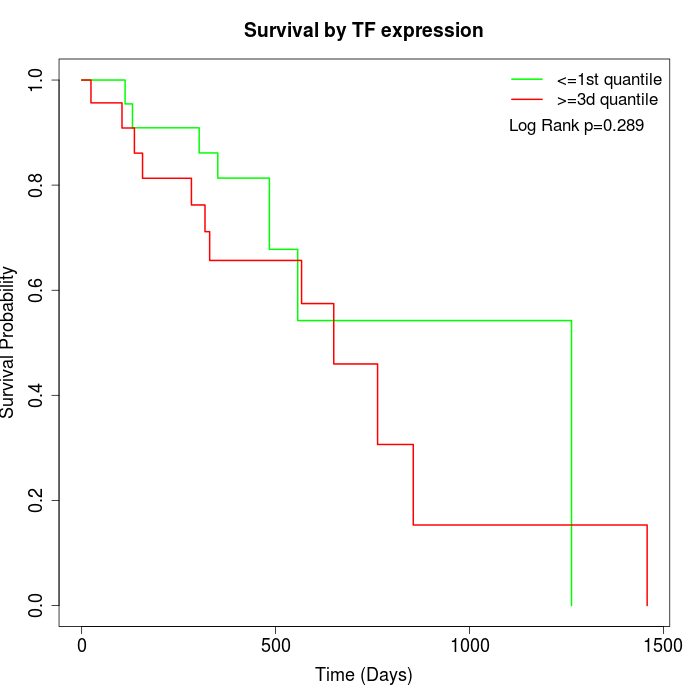
Survival by TF expression:
Note: Click image to view full size file.
Copy number change of TF:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TF | 7018 | 16 | 0 | 14 | |
| GSE20123 | TF | 7018 | 16 | 0 | 14 | |
| GSE43470 | TF | 7018 | 21 | 0 | 22 | |
| GSE46452 | TF | 7018 | 15 | 4 | 40 | |
| GSE47630 | TF | 7018 | 18 | 3 | 19 | |
| GSE54993 | TF | 7018 | 2 | 8 | 60 | |
| GSE54994 | TF | 7018 | 32 | 1 | 20 | |
| GSE60625 | TF | 7018 | 0 | 6 | 5 | |
| GSE74703 | TF | 7018 | 18 | 0 | 18 | |
| GSE74704 | TF | 7018 | 11 | 0 | 9 | |
| TCGA | TF | 7018 | 59 | 5 | 32 |
Total number of gains: 208; Total number of losses: 27; Total Number of normals: 253.
Somatic mutations of TF:
Generating mutation plots.
Highly correlated genes for TF:
Showing top 20/1003 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TF | CASC3 | 0.840785 | 3 | 0 | 3 |
| TF | LYNX1 | 0.807711 | 4 | 0 | 4 |
| TF | TEX101 | 0.773586 | 4 | 0 | 4 |
| TF | CNFN | 0.765754 | 4 | 0 | 4 |
| TF | MORN3 | 0.764087 | 4 | 0 | 3 |
| TF | CA13 | 0.750218 | 3 | 0 | 3 |
| TF | IQCF1 | 0.740138 | 4 | 0 | 3 |
| TF | ST6GALNAC1 | 0.72989 | 6 | 0 | 6 |
| TF | CYSRT1 | 0.728561 | 5 | 0 | 4 |
| TF | PRSS27 | 0.728364 | 5 | 0 | 4 |
| TF | SPATA31E1 | 0.72587 | 3 | 0 | 3 |
| TF | MPZL3 | 0.724921 | 6 | 0 | 6 |
| TF | NIPAL1 | 0.722545 | 3 | 0 | 3 |
| TF | SLC10A5 | 0.72214 | 3 | 0 | 3 |
| TF | THAP2 | 0.719861 | 3 | 0 | 3 |
| TF | CCDC7 | 0.719137 | 4 | 0 | 4 |
| TF | CXCL17 | 0.718777 | 7 | 0 | 7 |
| TF | MUC20 | 0.718112 | 6 | 0 | 6 |
| TF | GRPEL2 | 0.717611 | 5 | 0 | 5 |
| TF | RBM20 | 0.715674 | 5 | 0 | 5 |
For details and further investigation, click here