| Full name: Ly6/neurotoxin 1 | Alias Symbol: SLURP2 | ||
| Type: protein-coding gene | Cytoband: 8q24.3 | ||
| Entrez ID: 66004 | HGNC ID: HGNC:29604 | Ensembl Gene: ENSG00000180155 | OMIM ID: 606110 |
| Drug and gene relationship at DGIdb | |||
Expression of LYNX1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LYNX1 | 66004 | 1554179_s_at | -3.0364 | 0.1323 | |
| GSE26886 | LYNX1 | 66004 | 1554179_s_at | -3.7670 | 0.0000 | |
| GSE45670 | LYNX1 | 66004 | 1554179_s_at | -0.5737 | 0.3578 | |
| GSE53622 | LYNX1 | 66004 | 37830 | -2.6104 | 0.0000 | |
| GSE53624 | LYNX1 | 66004 | 37830 | -2.4363 | 0.0000 | |
| GSE63941 | LYNX1 | 66004 | 1554179_s_at | 0.5660 | 0.5673 | |
| GSE77861 | LYNX1 | 66004 | 1554179_s_at | -2.5356 | 0.0128 | |
| GSE97050 | LYNX1 | 66004 | A_33_P3418194 | -1.0630 | 0.1042 | |
| SRP007169 | LYNX1 | 66004 | RNAseq | -1.0092 | 0.1899 | |
| SRP064894 | LYNX1 | 66004 | RNAseq | -0.8192 | 0.0264 | |
| SRP133303 | LYNX1 | 66004 | RNAseq | -1.2466 | 0.0012 | |
| SRP159526 | LYNX1 | 66004 | RNAseq | -1.0386 | 0.0524 | |
| SRP193095 | LYNX1 | 66004 | RNAseq | -0.6586 | 0.0112 | |
| SRP219564 | LYNX1 | 66004 | RNAseq | -0.8852 | 0.2198 | |
| TCGA | LYNX1 | 66004 | RNAseq | -0.1076 | 0.6135 |
Upregulated datasets: 0; Downregulated datasets: 5.
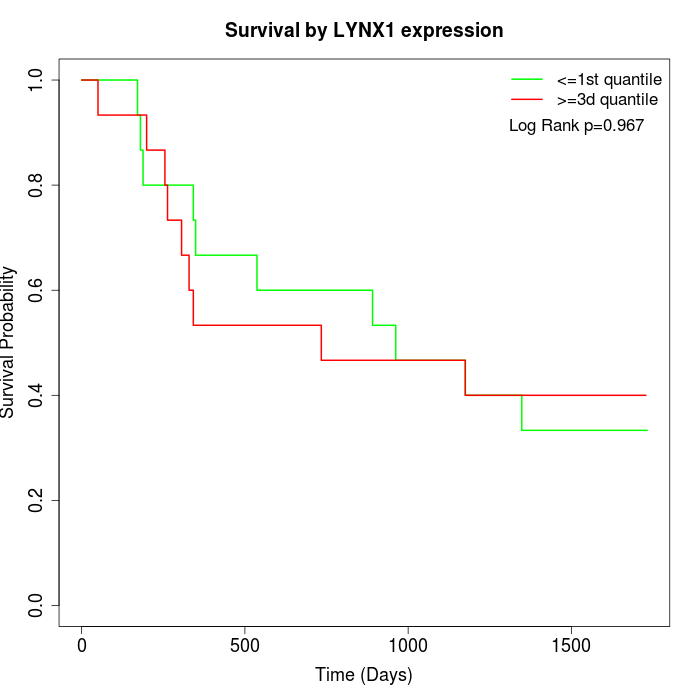
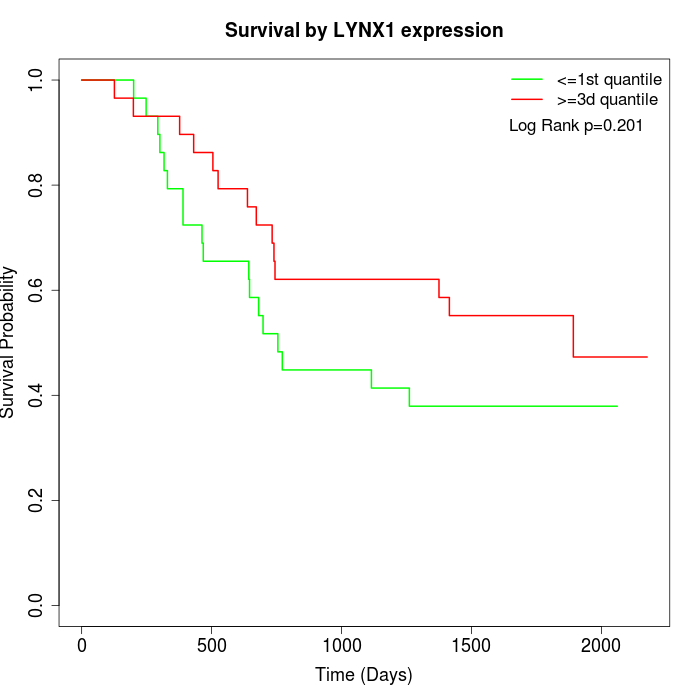
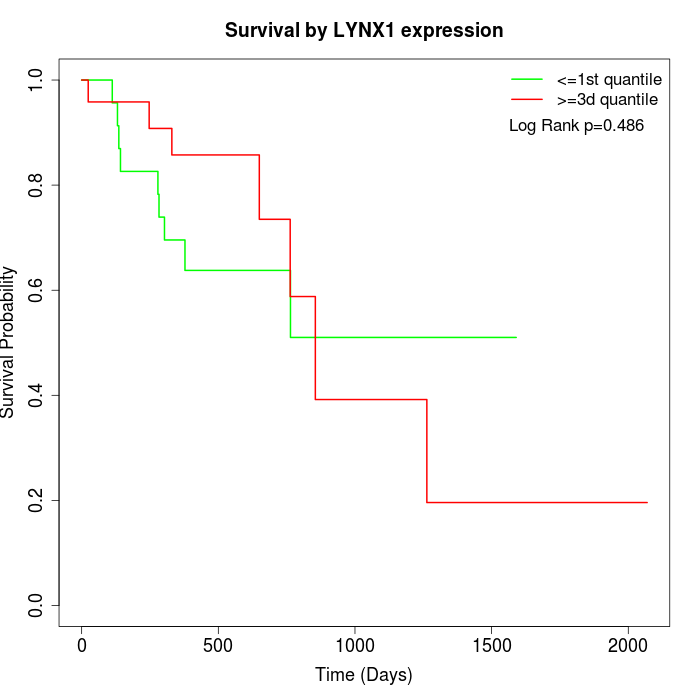
Survival by LYNX1 expression:
Note: Click image to view full size file.
Copy number change of LYNX1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LYNX1 | 66004 | 16 | 1 | 13 | |
| GSE20123 | LYNX1 | 66004 | 17 | 1 | 12 | |
| GSE43470 | LYNX1 | 66004 | 17 | 2 | 24 | |
| GSE46452 | LYNX1 | 66004 | 27 | 0 | 32 | |
| GSE47630 | LYNX1 | 66004 | 23 | 1 | 16 | |
| GSE54993 | LYNX1 | 66004 | 0 | 25 | 45 | |
| GSE54994 | LYNX1 | 66004 | 39 | 1 | 13 | |
| GSE60625 | LYNX1 | 66004 | 3 | 4 | 4 | |
| GSE74703 | LYNX1 | 66004 | 15 | 1 | 20 | |
| GSE74704 | LYNX1 | 66004 | 10 | 0 | 10 | |
| TCGA | LYNX1 | 66004 | 63 | 2 | 31 |
Total number of gains: 230; Total number of losses: 38; Total Number of normals: 220.
Somatic mutations of LYNX1:
Generating mutation plots.
Highly correlated genes for LYNX1:
Showing top 20/1495 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LYNX1 | CYSRT1 | 0.917377 | 6 | 0 | 6 |
| LYNX1 | VSIG10L | 0.89848 | 7 | 0 | 7 |
| LYNX1 | PRSS27 | 0.891701 | 8 | 0 | 8 |
| LYNX1 | SLURP1 | 0.875582 | 9 | 0 | 9 |
| LYNX1 | SPINK5 | 0.873375 | 8 | 0 | 8 |
| LYNX1 | TGM3 | 0.869835 | 8 | 0 | 8 |
| LYNX1 | SPINK7 | 0.86816 | 8 | 0 | 8 |
| LYNX1 | CLIC3 | 0.868156 | 6 | 0 | 6 |
| LYNX1 | CNFN | 0.867591 | 8 | 0 | 8 |
| LYNX1 | LYPD2 | 0.866207 | 3 | 0 | 3 |
| LYNX1 | NCCRP1 | 0.863043 | 4 | 0 | 4 |
| LYNX1 | PPL | 0.857995 | 8 | 0 | 8 |
| LYNX1 | CRNN | 0.856614 | 8 | 0 | 8 |
| LYNX1 | KLK13 | 0.85094 | 9 | 0 | 9 |
| LYNX1 | IL11RA | 0.850348 | 3 | 0 | 3 |
| LYNX1 | TMPRSS11B | 0.848371 | 8 | 0 | 8 |
| LYNX1 | SPRR3 | 0.844142 | 8 | 0 | 8 |
| LYNX1 | CSTB | 0.843595 | 8 | 0 | 8 |
| LYNX1 | CAPN14 | 0.843125 | 7 | 0 | 7 |
| LYNX1 | ECM1 | 0.833116 | 8 | 0 | 8 |
For details and further investigation, click here