| Full name: NIPA like domain containing 1 | Alias Symbol: DKFZp686A06115 | ||
| Type: protein-coding gene | Cytoband: 4p12 | ||
| Entrez ID: 152519 | HGNC ID: HGNC:27194 | Ensembl Gene: ENSG00000163293 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of NIPAL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | NIPAL1 | 152519 | 41319 | -1.1088 | 0.0000 | |
| GSE53624 | NIPAL1 | 152519 | 41319 | -1.2399 | 0.0000 | |
| GSE97050 | NIPAL1 | 152519 | A_33_P3417086 | -0.1518 | 0.8415 | |
| SRP007169 | NIPAL1 | 152519 | RNAseq | -3.6350 | 0.0000 | |
| SRP008496 | NIPAL1 | 152519 | RNAseq | -3.4023 | 0.0000 | |
| SRP064894 | NIPAL1 | 152519 | RNAseq | -2.7915 | 0.0000 | |
| SRP133303 | NIPAL1 | 152519 | RNAseq | -2.0513 | 0.0000 | |
| SRP159526 | NIPAL1 | 152519 | RNAseq | -2.9549 | 0.0000 | |
| SRP193095 | NIPAL1 | 152519 | RNAseq | -2.3426 | 0.0000 | |
| SRP219564 | NIPAL1 | 152519 | RNAseq | -2.9763 | 0.0032 | |
| TCGA | NIPAL1 | 152519 | RNAseq | -0.0302 | 0.8805 |
Upregulated datasets: 0; Downregulated datasets: 9.
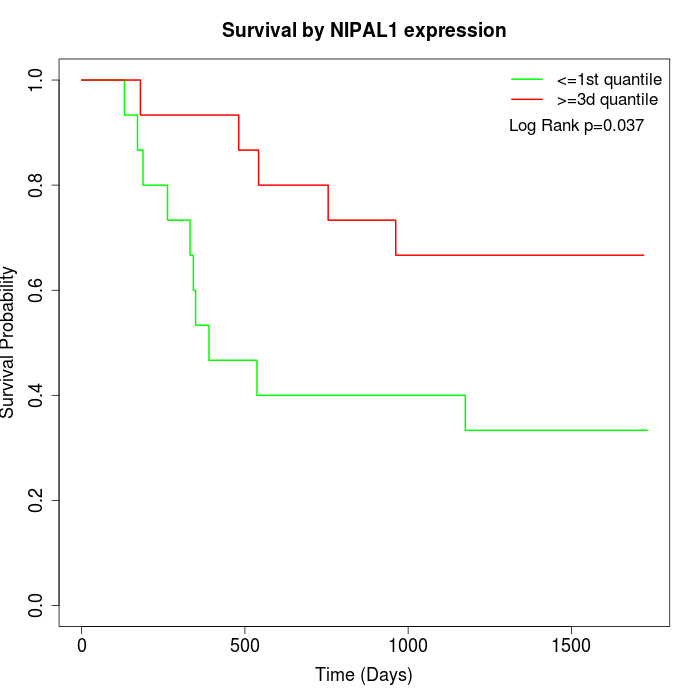
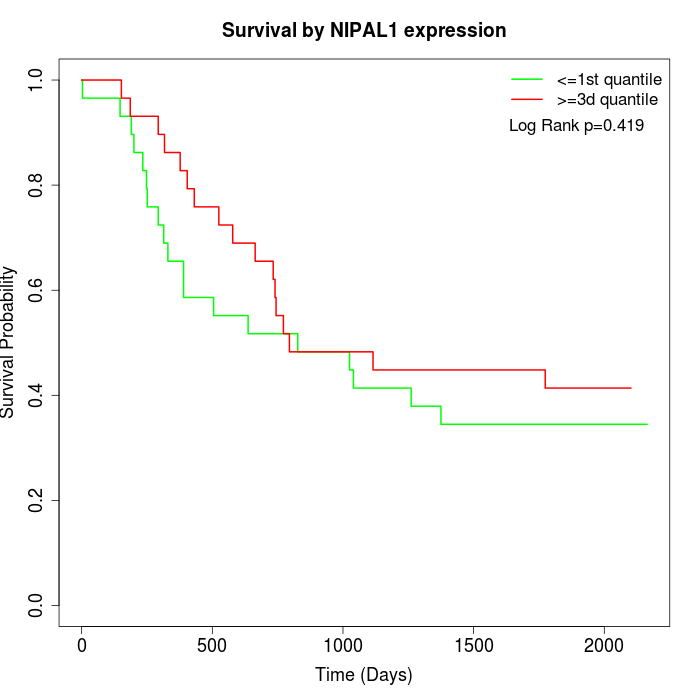
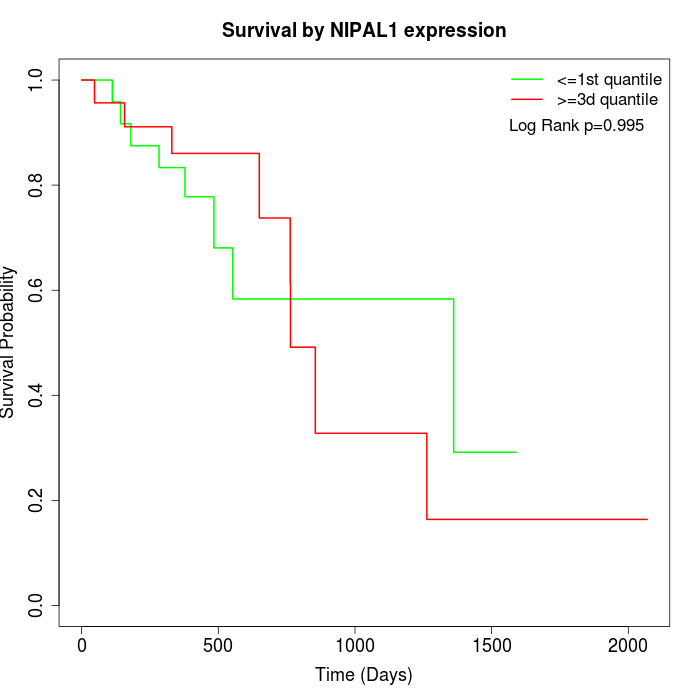
Survival by NIPAL1 expression:
Note: Click image to view full size file.
Copy number change of NIPAL1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NIPAL1 | 152519 | 1 | 15 | 14 | |
| GSE20123 | NIPAL1 | 152519 | 1 | 15 | 14 | |
| GSE43470 | NIPAL1 | 152519 | 0 | 13 | 30 | |
| GSE46452 | NIPAL1 | 152519 | 1 | 37 | 21 | |
| GSE47630 | NIPAL1 | 152519 | 1 | 18 | 21 | |
| GSE54993 | NIPAL1 | 152519 | 7 | 0 | 63 | |
| GSE54994 | NIPAL1 | 152519 | 4 | 9 | 40 | |
| GSE60625 | NIPAL1 | 152519 | 0 | 0 | 11 | |
| GSE74703 | NIPAL1 | 152519 | 0 | 11 | 25 | |
| GSE74704 | NIPAL1 | 152519 | 0 | 9 | 11 | |
| TCGA | NIPAL1 | 152519 | 18 | 30 | 48 |
Total number of gains: 33; Total number of losses: 157; Total Number of normals: 298.
Somatic mutations of NIPAL1:
Generating mutation plots.
Highly correlated genes for NIPAL1:
Showing top 20/539 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NIPAL1 | RAB25 | 0.862083 | 3 | 0 | 3 |
| NIPAL1 | ARHGAP27 | 0.853031 | 3 | 0 | 3 |
| NIPAL1 | ZDHHC13 | 0.85001 | 3 | 0 | 3 |
| NIPAL1 | SYTL1 | 0.848417 | 3 | 0 | 3 |
| NIPAL1 | MPZL3 | 0.847335 | 3 | 0 | 3 |
| NIPAL1 | DUOX1 | 0.837372 | 3 | 0 | 3 |
| NIPAL1 | NMU | 0.836241 | 3 | 0 | 3 |
| NIPAL1 | YOD1 | 0.833843 | 3 | 0 | 3 |
| NIPAL1 | DENND2C | 0.831238 | 3 | 0 | 3 |
| NIPAL1 | TMEM40 | 0.829137 | 3 | 0 | 3 |
| NIPAL1 | FUT3 | 0.826233 | 3 | 0 | 3 |
| NIPAL1 | FAM160A1 | 0.825099 | 3 | 0 | 3 |
| NIPAL1 | MPZL2 | 0.822461 | 3 | 0 | 3 |
| NIPAL1 | NUAK2 | 0.822381 | 3 | 0 | 3 |
| NIPAL1 | BARX2 | 0.821041 | 3 | 0 | 3 |
| NIPAL1 | PPP1R13L | 0.818371 | 3 | 0 | 3 |
| NIPAL1 | TIAM1 | 0.81405 | 3 | 0 | 3 |
| NIPAL1 | ATG9B | 0.813638 | 3 | 0 | 3 |
| NIPAL1 | EXPH5 | 0.80869 | 3 | 0 | 3 |
| NIPAL1 | DGKA | 0.805661 | 3 | 0 | 3 |
For details and further investigation, click here