| Full name: troponin C2, fast skeletal type | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 20q13.12 | ||
| Entrez ID: 7125 | HGNC ID: HGNC:11944 | Ensembl Gene: ENSG00000101470 | OMIM ID: 191039 |
| Drug and gene relationship at DGIdb | |||
TNNC2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04020 | Calcium signaling pathway |
Expression of TNNC2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TNNC2 | 7125 | 205388_at | -0.0016 | 0.9985 | |
| GSE20347 | TNNC2 | 7125 | 205388_at | -0.0291 | 0.6773 | |
| GSE23400 | TNNC2 | 7125 | 205388_at | -0.1049 | 0.0061 | |
| GSE26886 | TNNC2 | 7125 | 205388_at | 0.2663 | 0.0408 | |
| GSE29001 | TNNC2 | 7125 | 205388_at | -0.0309 | 0.8904 | |
| GSE38129 | TNNC2 | 7125 | 205388_at | -0.2804 | 0.0055 | |
| GSE45670 | TNNC2 | 7125 | 205388_at | -0.4123 | 0.0000 | |
| GSE63941 | TNNC2 | 7125 | 205388_at | -0.0456 | 0.8925 | |
| GSE77861 | TNNC2 | 7125 | 205388_at | -0.0145 | 0.9390 | |
| GSE97050 | TNNC2 | 7125 | A_23_P131825 | -0.2896 | 0.2466 | |
| SRP219564 | TNNC2 | 7125 | RNAseq | -1.4817 | 0.0580 | |
| TCGA | TNNC2 | 7125 | RNAseq | -0.6122 | 0.4109 |
Upregulated datasets: 0; Downregulated datasets: 0.
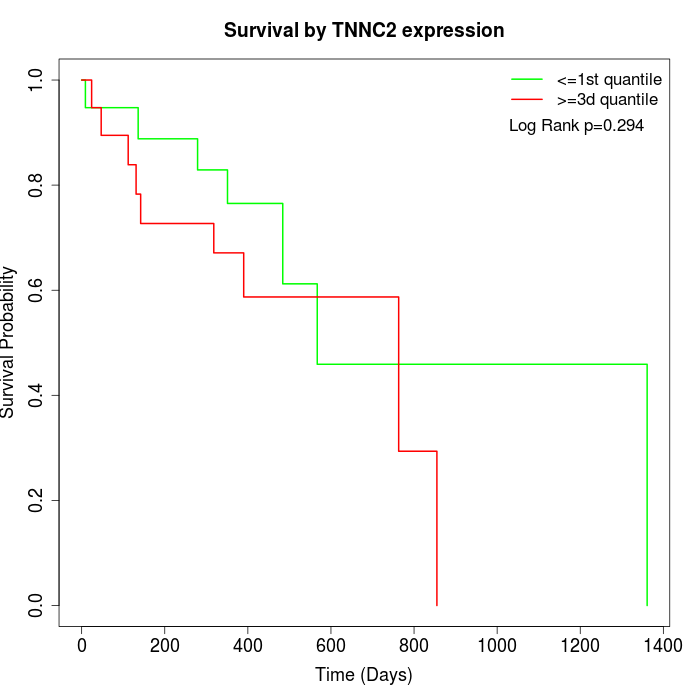
Survival by TNNC2 expression:
Note: Click image to view full size file.
Copy number change of TNNC2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TNNC2 | 7125 | 14 | 1 | 15 | |
| GSE20123 | TNNC2 | 7125 | 14 | 1 | 15 | |
| GSE43470 | TNNC2 | 7125 | 12 | 1 | 30 | |
| GSE46452 | TNNC2 | 7125 | 29 | 0 | 30 | |
| GSE47630 | TNNC2 | 7125 | 24 | 1 | 15 | |
| GSE54993 | TNNC2 | 7125 | 0 | 18 | 52 | |
| GSE54994 | TNNC2 | 7125 | 25 | 0 | 28 | |
| GSE60625 | TNNC2 | 7125 | 0 | 0 | 11 | |
| GSE74703 | TNNC2 | 7125 | 10 | 1 | 25 | |
| GSE74704 | TNNC2 | 7125 | 10 | 0 | 10 | |
| TCGA | TNNC2 | 7125 | 46 | 3 | 47 |
Total number of gains: 184; Total number of losses: 26; Total Number of normals: 278.
Somatic mutations of TNNC2:
Generating mutation plots.
Highly correlated genes for TNNC2:
Showing top 20/311 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TNNC2 | RASL10B | 0.793846 | 3 | 0 | 3 |
| TNNC2 | C19orf44 | 0.77057 | 3 | 0 | 3 |
| TNNC2 | CPAMD8 | 0.768703 | 3 | 0 | 3 |
| TNNC2 | ZNF524 | 0.755599 | 3 | 0 | 3 |
| TNNC2 | PICK1 | 0.753399 | 4 | 0 | 4 |
| TNNC2 | SPATA2L | 0.747373 | 3 | 0 | 3 |
| TNNC2 | ZNF182 | 0.743194 | 3 | 0 | 3 |
| TNNC2 | ZBTB45 | 0.728826 | 3 | 0 | 3 |
| TNNC2 | FMR1NB | 0.727647 | 3 | 0 | 3 |
| TNNC2 | NPBWR1 | 0.724818 | 3 | 0 | 3 |
| TNNC2 | HDAC10 | 0.721644 | 3 | 0 | 3 |
| TNNC2 | C20orf141 | 0.719013 | 4 | 0 | 3 |
| TNNC2 | NRSN1 | 0.716118 | 3 | 0 | 3 |
| TNNC2 | TBX21 | 0.715046 | 3 | 0 | 3 |
| TNNC2 | RCOR2 | 0.713487 | 3 | 0 | 3 |
| TNNC2 | ATP7B | 0.711811 | 3 | 0 | 3 |
| TNNC2 | C19orf53 | 0.711792 | 3 | 0 | 3 |
| TNNC2 | LIM2 | 0.710801 | 4 | 0 | 4 |
| TNNC2 | GAS6-AS1 | 0.709889 | 3 | 0 | 3 |
| TNNC2 | ADAMTSL3 | 0.706296 | 3 | 0 | 3 |
For details and further investigation, click here