| Full name: tRNA isopentenyltransferase 1 | Alias Symbol: FLJ20061|IPT | ||
| Type: protein-coding gene | Cytoband: 1p34.2 | ||
| Entrez ID: 54802 | HGNC ID: HGNC:20286 | Ensembl Gene: ENSG00000043514 | OMIM ID: 617840 |
| Drug and gene relationship at DGIdb | |||
Expression of TRIT1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIT1 | 54802 | 218617_at | 0.2795 | 0.5330 | |
| GSE20347 | TRIT1 | 54802 | 218617_at | 0.1091 | 0.3279 | |
| GSE23400 | TRIT1 | 54802 | 218617_at | 0.4450 | 0.0006 | |
| GSE26886 | TRIT1 | 54802 | 218617_at | 0.3169 | 0.1909 | |
| GSE29001 | TRIT1 | 54802 | 218617_at | 0.2451 | 0.5768 | |
| GSE38129 | TRIT1 | 54802 | 218617_at | 0.3396 | 0.0046 | |
| GSE45670 | TRIT1 | 54802 | 218617_at | 0.0015 | 0.9908 | |
| GSE53622 | TRIT1 | 54802 | 138962 | 0.2851 | 0.0000 | |
| GSE53624 | TRIT1 | 54802 | 138962 | 0.3340 | 0.0000 | |
| GSE63941 | TRIT1 | 54802 | 218617_at | 0.7357 | 0.0967 | |
| GSE77861 | TRIT1 | 54802 | 218617_at | 0.2206 | 0.5419 | |
| GSE97050 | TRIT1 | 54802 | A_23_P12062 | -0.1637 | 0.5885 | |
| SRP007169 | TRIT1 | 54802 | RNAseq | 0.1902 | 0.6396 | |
| SRP008496 | TRIT1 | 54802 | RNAseq | 0.0446 | 0.8869 | |
| SRP064894 | TRIT1 | 54802 | RNAseq | 0.2213 | 0.1729 | |
| SRP133303 | TRIT1 | 54802 | RNAseq | 0.0861 | 0.5249 | |
| SRP159526 | TRIT1 | 54802 | RNAseq | 1.4618 | 0.0154 | |
| SRP193095 | TRIT1 | 54802 | RNAseq | -0.0306 | 0.8473 | |
| SRP219564 | TRIT1 | 54802 | RNAseq | 0.3288 | 0.4284 | |
| TCGA | TRIT1 | 54802 | RNAseq | 0.2860 | 0.0046 |
Upregulated datasets: 1; Downregulated datasets: 0.
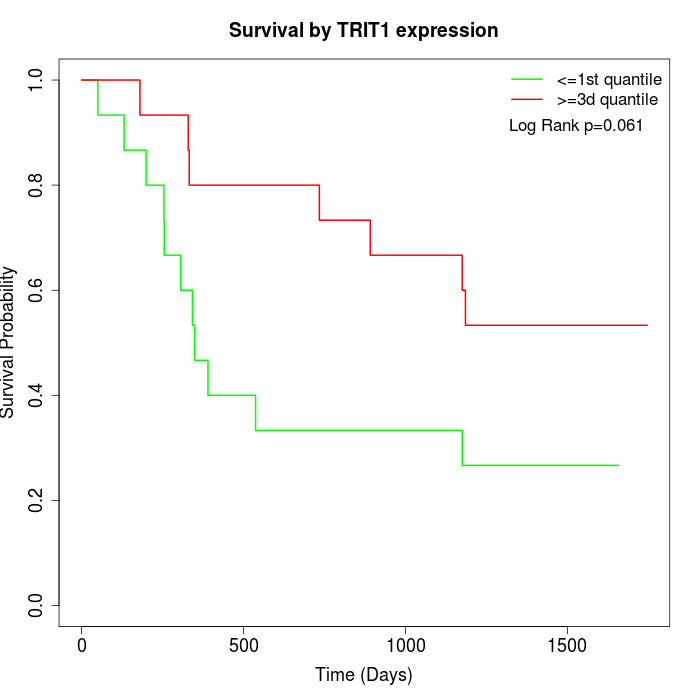
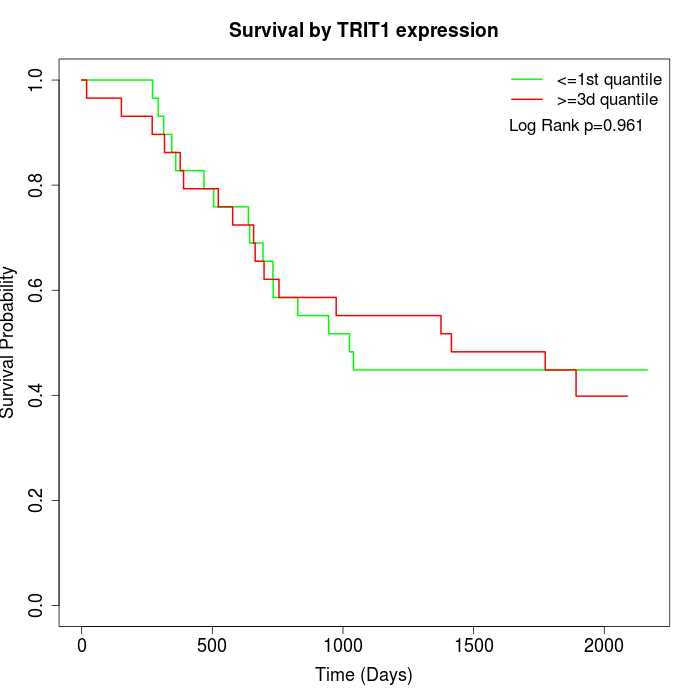
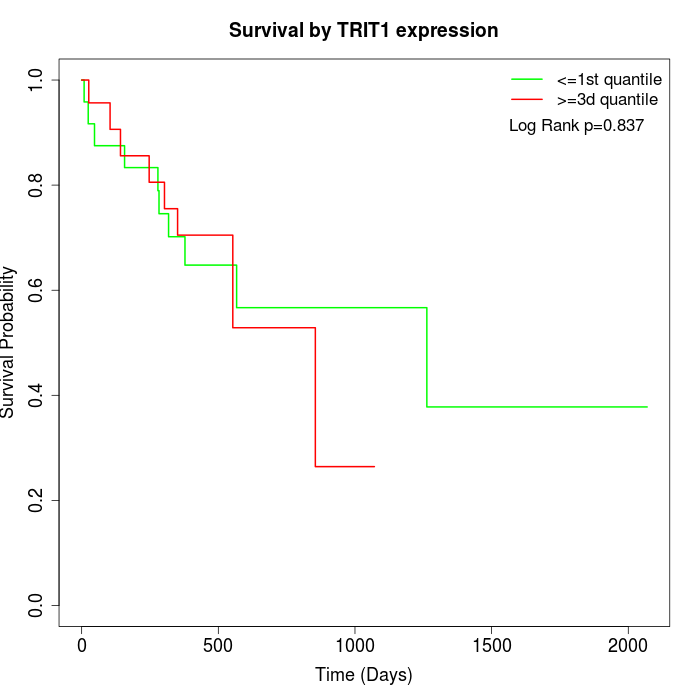
Survival by TRIT1 expression:
Note: Click image to view full size file.
Copy number change of TRIT1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIT1 | 54802 | 3 | 4 | 23 | |
| GSE20123 | TRIT1 | 54802 | 2 | 3 | 25 | |
| GSE43470 | TRIT1 | 54802 | 7 | 2 | 34 | |
| GSE46452 | TRIT1 | 54802 | 4 | 1 | 54 | |
| GSE47630 | TRIT1 | 54802 | 9 | 2 | 29 | |
| GSE54993 | TRIT1 | 54802 | 0 | 1 | 69 | |
| GSE54994 | TRIT1 | 54802 | 13 | 2 | 38 | |
| GSE60625 | TRIT1 | 54802 | 0 | 0 | 11 | |
| GSE74703 | TRIT1 | 54802 | 6 | 1 | 29 | |
| GSE74704 | TRIT1 | 54802 | 1 | 0 | 19 | |
| TCGA | TRIT1 | 54802 | 16 | 16 | 64 |
Total number of gains: 61; Total number of losses: 32; Total Number of normals: 395.
Somatic mutations of TRIT1:
Generating mutation plots.
Highly correlated genes for TRIT1:
Showing top 20/530 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIT1 | PPIL4 | 0.798875 | 4 | 0 | 4 |
| TRIT1 | MTF1 | 0.788697 | 3 | 0 | 3 |
| TRIT1 | PPIL1 | 0.78318 | 3 | 0 | 3 |
| TRIT1 | MED26 | 0.777106 | 3 | 0 | 3 |
| TRIT1 | RRAGC | 0.771018 | 3 | 0 | 3 |
| TRIT1 | SLC38A7 | 0.759887 | 3 | 0 | 3 |
| TRIT1 | TRIAP1 | 0.753403 | 4 | 0 | 4 |
| TRIT1 | ZNF764 | 0.747609 | 3 | 0 | 3 |
| TRIT1 | GORAB | 0.747264 | 3 | 0 | 3 |
| TRIT1 | CCNJ | 0.747083 | 3 | 0 | 3 |
| TRIT1 | RRP36 | 0.739204 | 3 | 0 | 3 |
| TRIT1 | PPP2R3C | 0.725927 | 3 | 0 | 3 |
| TRIT1 | BTF3L4 | 0.725598 | 4 | 0 | 4 |
| TRIT1 | ZNF121 | 0.720381 | 4 | 0 | 4 |
| TRIT1 | QSER1 | 0.713835 | 3 | 0 | 3 |
| TRIT1 | TRUB1 | 0.709276 | 3 | 0 | 3 |
| TRIT1 | USP34 | 0.708378 | 4 | 0 | 3 |
| TRIT1 | PSMG3 | 0.704669 | 3 | 0 | 3 |
| TRIT1 | ARTN | 0.703417 | 3 | 0 | 3 |
| TRIT1 | ZNF227 | 0.696215 | 4 | 0 | 3 |
For details and further investigation, click here