| Full name: ATP binding cassette subfamily G member 5 | Alias Symbol: STSL | ||
| Type: protein-coding gene | Cytoband: 2p21 | ||
| Entrez ID: 64240 | HGNC ID: HGNC:13886 | Ensembl Gene: ENSG00000138075 | OMIM ID: 605459 |
| Drug and gene relationship at DGIdb | |||
Expression of ABCG5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ABCG5 | 64240 | 220383_at | 0.0632 | 0.8360 | |
| GSE20347 | ABCG5 | 64240 | 220383_at | -0.1932 | 0.0029 | |
| GSE23400 | ABCG5 | 64240 | 220383_at | -0.1418 | 0.0001 | |
| GSE26886 | ABCG5 | 64240 | 220383_at | 0.0726 | 0.4825 | |
| GSE29001 | ABCG5 | 64240 | 220383_at | -0.2126 | 0.1974 | |
| GSE38129 | ABCG5 | 64240 | 220383_at | -0.1886 | 0.0027 | |
| GSE45670 | ABCG5 | 64240 | 220383_at | 0.0420 | 0.7988 | |
| GSE53622 | ABCG5 | 64240 | 27041 | 0.0823 | 0.4527 | |
| GSE53624 | ABCG5 | 64240 | 27041 | 0.0786 | 0.4022 | |
| GSE63941 | ABCG5 | 64240 | 220383_at | -0.0224 | 0.9195 | |
| GSE77861 | ABCG5 | 64240 | 220383_at | -0.0968 | 0.5383 | |
| TCGA | ABCG5 | 64240 | RNAseq | 1.0846 | 0.0735 |
Upregulated datasets: 0; Downregulated datasets: 0.
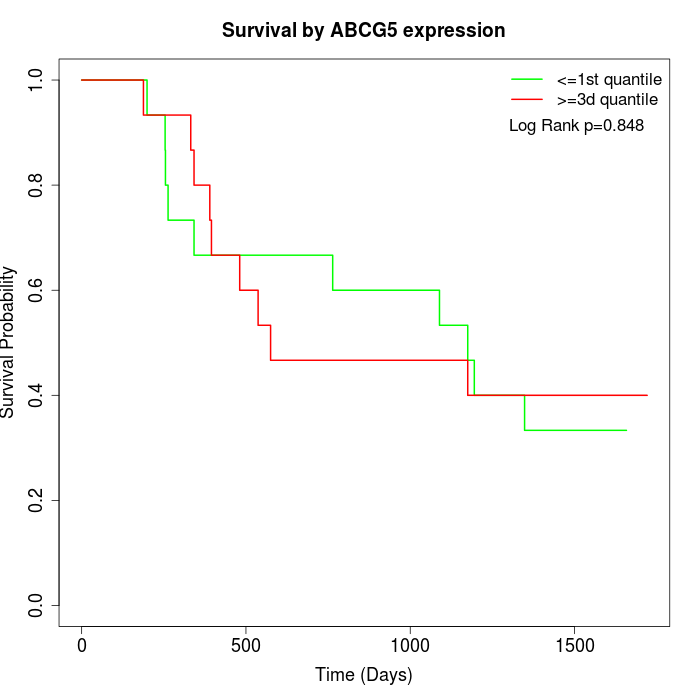
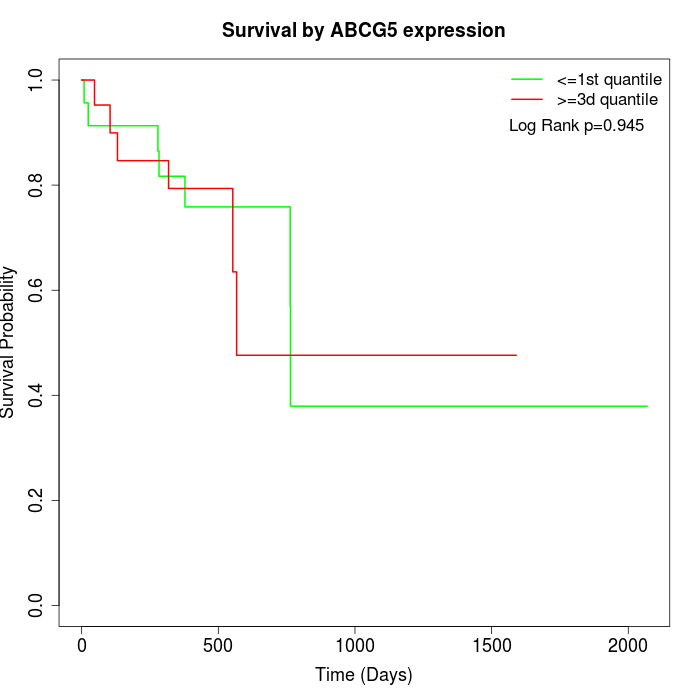
Survival by ABCG5 expression:
Note: Click image to view full size file.
Copy number change of ABCG5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ABCG5 | 64240 | 10 | 1 | 19 | |
| GSE20123 | ABCG5 | 64240 | 9 | 1 | 20 | |
| GSE43470 | ABCG5 | 64240 | 4 | 0 | 39 | |
| GSE46452 | ABCG5 | 64240 | 1 | 4 | 54 | |
| GSE47630 | ABCG5 | 64240 | 7 | 0 | 33 | |
| GSE54993 | ABCG5 | 64240 | 0 | 5 | 65 | |
| GSE54994 | ABCG5 | 64240 | 10 | 0 | 43 | |
| GSE60625 | ABCG5 | 64240 | 0 | 3 | 8 | |
| GSE74703 | ABCG5 | 64240 | 4 | 0 | 32 | |
| GSE74704 | ABCG5 | 64240 | 9 | 1 | 10 | |
| TCGA | ABCG5 | 64240 | 36 | 2 | 58 |
Total number of gains: 90; Total number of losses: 17; Total Number of normals: 381.
Somatic mutations of ABCG5:
Generating mutation plots.
Highly correlated genes for ABCG5:
Showing top 20/194 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ABCG5 | DRP2 | 0.686806 | 3 | 0 | 3 |
| ABCG5 | ACSBG2 | 0.655754 | 4 | 0 | 3 |
| ABCG5 | VPS53 | 0.645087 | 3 | 0 | 3 |
| ABCG5 | PDE6A | 0.643477 | 4 | 0 | 4 |
| ABCG5 | THPO | 0.634069 | 3 | 0 | 3 |
| ABCG5 | MEP1B | 0.628932 | 5 | 0 | 5 |
| ABCG5 | CPS1-IT1 | 0.626292 | 4 | 0 | 4 |
| ABCG5 | USH1C | 0.619656 | 4 | 0 | 3 |
| ABCG5 | CYLC1 | 0.618678 | 4 | 0 | 3 |
| ABCG5 | GRPR | 0.61685 | 5 | 0 | 3 |
| ABCG5 | CCR4 | 0.616226 | 3 | 0 | 3 |
| ABCG5 | MC5R | 0.616129 | 5 | 0 | 4 |
| ABCG5 | CEACAM3 | 0.607136 | 4 | 0 | 3 |
| ABCG5 | ART1 | 0.606975 | 4 | 0 | 3 |
| ABCG5 | CT55 | 0.603008 | 3 | 0 | 3 |
| ABCG5 | RARG | 0.601255 | 5 | 0 | 4 |
| ABCG5 | B3GALT1 | 0.596956 | 3 | 0 | 3 |
| ABCG5 | KCNQ1DN | 0.594304 | 5 | 0 | 5 |
| ABCG5 | SPIN2A | 0.593162 | 5 | 0 | 4 |
| ABCG5 | GYPA | 0.592354 | 3 | 0 | 3 |
For details and further investigation, click here