| Full name: beta-1,3-galactosyltransferase 1 | Alias Symbol: beta3Gal-T1 | ||
| Type: protein-coding gene | Cytoband: 2q24.3 | ||
| Entrez ID: 8708 | HGNC ID: HGNC:916 | Ensembl Gene: ENSG00000172318 | OMIM ID: 603093 |
| Drug and gene relationship at DGIdb | |||
Expression of B3GALT1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | B3GALT1 | 8708 | 220829_s_at | 0.1503 | 0.5653 | |
| GSE20347 | B3GALT1 | 8708 | 220829_s_at | 0.0436 | 0.5311 | |
| GSE23400 | B3GALT1 | 8708 | 220829_s_at | -0.0387 | 0.2597 | |
| GSE26886 | B3GALT1 | 8708 | 222969_at | -0.1077 | 0.2025 | |
| GSE29001 | B3GALT1 | 8708 | 220829_s_at | 0.0151 | 0.9252 | |
| GSE38129 | B3GALT1 | 8708 | 220829_s_at | 0.0101 | 0.8866 | |
| GSE45670 | B3GALT1 | 8708 | 222969_at | -0.0334 | 0.6820 | |
| GSE63941 | B3GALT1 | 8708 | 222969_at | -0.0723 | 0.6594 | |
| GSE77861 | B3GALT1 | 8708 | 222969_at | -0.0901 | 0.3649 | |
| GSE97050 | B3GALT1 | 8708 | A_23_P108564 | 0.1379 | 0.4950 | |
| TCGA | B3GALT1 | 8708 | RNAseq | -0.4105 | 0.5812 |
Upregulated datasets: 0; Downregulated datasets: 0.
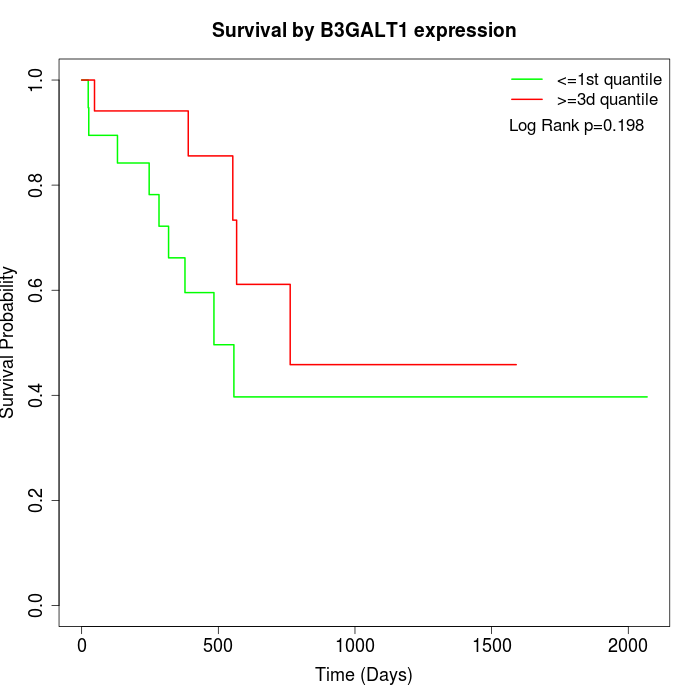
Survival by B3GALT1 expression:
Note: Click image to view full size file.
Copy number change of B3GALT1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | B3GALT1 | 8708 | 6 | 0 | 24 | |
| GSE20123 | B3GALT1 | 8708 | 6 | 0 | 24 | |
| GSE43470 | B3GALT1 | 8708 | 5 | 1 | 37 | |
| GSE46452 | B3GALT1 | 8708 | 1 | 4 | 54 | |
| GSE47630 | B3GALT1 | 8708 | 5 | 3 | 32 | |
| GSE54993 | B3GALT1 | 8708 | 0 | 5 | 65 | |
| GSE54994 | B3GALT1 | 8708 | 11 | 2 | 40 | |
| GSE60625 | B3GALT1 | 8708 | 0 | 3 | 8 | |
| GSE74703 | B3GALT1 | 8708 | 4 | 1 | 31 | |
| GSE74704 | B3GALT1 | 8708 | 3 | 0 | 17 | |
| TCGA | B3GALT1 | 8708 | 24 | 10 | 62 |
Total number of gains: 65; Total number of losses: 29; Total Number of normals: 394.
Somatic mutations of B3GALT1:
Generating mutation plots.
Highly correlated genes for B3GALT1:
Showing top 20/486 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| B3GALT1 | TBX4 | 0.835707 | 3 | 0 | 3 |
| B3GALT1 | ABCB9 | 0.779911 | 3 | 0 | 3 |
| B3GALT1 | LIMD1 | 0.770434 | 3 | 0 | 3 |
| B3GALT1 | RAG2 | 0.758568 | 3 | 0 | 3 |
| B3GALT1 | AVPR2 | 0.758336 | 3 | 0 | 3 |
| B3GALT1 | CPLX2 | 0.743667 | 3 | 0 | 3 |
| B3GALT1 | PNMA3 | 0.736457 | 3 | 0 | 3 |
| B3GALT1 | FCN2 | 0.735107 | 6 | 0 | 6 |
| B3GALT1 | THAP3 | 0.734276 | 3 | 0 | 3 |
| B3GALT1 | CADPS | 0.733173 | 3 | 0 | 3 |
| B3GALT1 | CRYGC | 0.72849 | 3 | 0 | 3 |
| B3GALT1 | CA6 | 0.726531 | 3 | 0 | 3 |
| B3GALT1 | DAB1 | 0.724726 | 4 | 0 | 3 |
| B3GALT1 | ADRA1A | 0.722232 | 4 | 0 | 3 |
| B3GALT1 | CDH22 | 0.721533 | 3 | 0 | 3 |
| B3GALT1 | LY6G5C | 0.721271 | 3 | 0 | 3 |
| B3GALT1 | OLIG3 | 0.72055 | 3 | 0 | 3 |
| B3GALT1 | HSD17B14 | 0.718772 | 3 | 0 | 3 |
| B3GALT1 | ABCC2 | 0.716834 | 3 | 0 | 3 |
| B3GALT1 | SPINK2 | 0.71514 | 3 | 0 | 3 |
For details and further investigation, click here