| Full name: glycophorin A (MNS blood group) | Alias Symbol: GPA|MN|CD235a | ||
| Type: protein-coding gene | Cytoband: 4q31.21 | ||
| Entrez ID: 2993 | HGNC ID: HGNC:4702 | Ensembl Gene: ENSG00000170180 | OMIM ID: 617922 |
| Drug and gene relationship at DGIdb | |||
Expression of GYPA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GYPA | 2993 | 205838_at | 0.2173 | 0.2440 | |
| GSE20347 | GYPA | 2993 | 211821_x_at | 0.0236 | 0.7383 | |
| GSE23400 | GYPA | 2993 | 205838_at | -0.0987 | 0.0013 | |
| GSE26886 | GYPA | 2993 | 211820_x_at | 0.1292 | 0.1959 | |
| GSE29001 | GYPA | 2993 | 211820_x_at | 0.0129 | 0.9393 | |
| GSE38129 | GYPA | 2993 | 211821_x_at | -0.0892 | 0.1931 | |
| GSE45670 | GYPA | 2993 | 211821_x_at | 0.0275 | 0.6973 | |
| GSE53622 | GYPA | 2993 | 93459 | 0.1307 | 0.5292 | |
| GSE53624 | GYPA | 2993 | 93459 | 0.4070 | 0.0445 | |
| GSE63941 | GYPA | 2993 | 205838_at | -0.0278 | 0.8716 | |
| GSE77861 | GYPA | 2993 | 211820_x_at | -0.0723 | 0.4171 | |
| GSE97050 | GYPA | 2993 | A_33_P3362933 | 0.2359 | 0.3958 | |
| SRP133303 | GYPA | 2993 | RNAseq | 0.0053 | 0.9843 |
Upregulated datasets: 0; Downregulated datasets: 0.
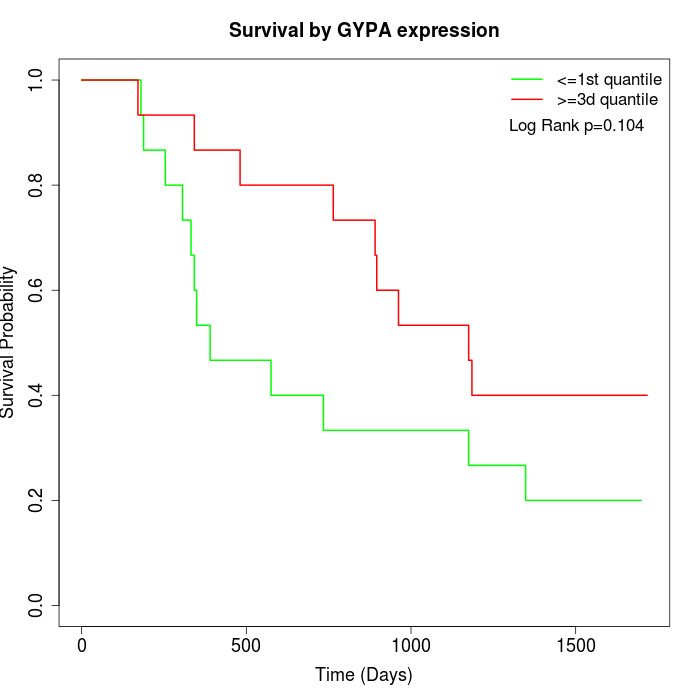
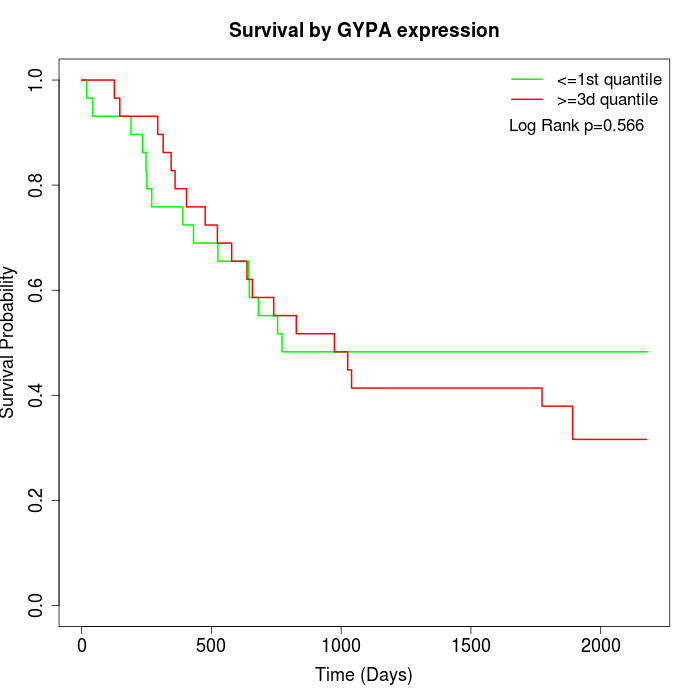
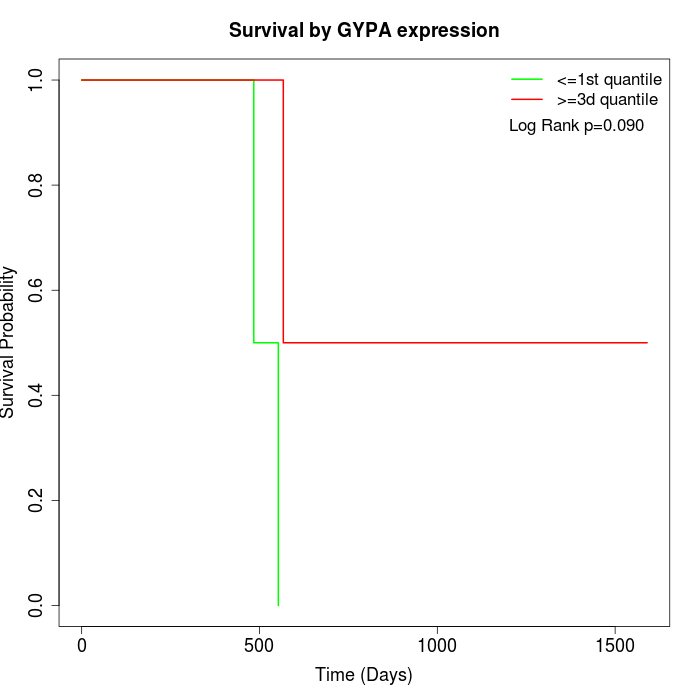
Survival by GYPA expression:
Note: Click image to view full size file.
Copy number change of GYPA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GYPA | 2993 | 0 | 12 | 18 | |
| GSE20123 | GYPA | 2993 | 0 | 12 | 18 | |
| GSE43470 | GYPA | 2993 | 0 | 13 | 30 | |
| GSE46452 | GYPA | 2993 | 1 | 36 | 22 | |
| GSE47630 | GYPA | 2993 | 0 | 22 | 18 | |
| GSE54993 | GYPA | 2993 | 10 | 0 | 60 | |
| GSE54994 | GYPA | 2993 | 2 | 11 | 40 | |
| GSE60625 | GYPA | 2993 | 0 | 1 | 10 | |
| GSE74703 | GYPA | 2993 | 0 | 11 | 25 | |
| GSE74704 | GYPA | 2993 | 0 | 6 | 14 | |
| TCGA | GYPA | 2993 | 13 | 30 | 53 |
Total number of gains: 26; Total number of losses: 154; Total Number of normals: 308.
Somatic mutations of GYPA:
Generating mutation plots.
Highly correlated genes for GYPA:
Showing top 20/851 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GYPA | NOL4 | 0.774049 | 3 | 0 | 3 |
| GYPA | GIT1 | 0.740249 | 3 | 0 | 3 |
| GYPA | KHDRBS2 | 0.720498 | 4 | 0 | 4 |
| GYPA | SCGN | 0.711708 | 3 | 0 | 3 |
| GYPA | ZNF460 | 0.704529 | 5 | 0 | 5 |
| GYPA | MTNR1B | 0.698978 | 4 | 0 | 4 |
| GYPA | FAM153A | 0.693004 | 5 | 0 | 5 |
| GYPA | SLC9A7 | 0.686477 | 4 | 0 | 4 |
| GYPA | EPHA5 | 0.684107 | 4 | 0 | 4 |
| GYPA | ZMAT4 | 0.675004 | 4 | 0 | 4 |
| GYPA | ECE2 | 0.673817 | 5 | 0 | 5 |
| GYPA | NOS2 | 0.671451 | 3 | 0 | 3 |
| GYPA | CPA2 | 0.670893 | 3 | 0 | 3 |
| GYPA | PEAK1 | 0.668626 | 4 | 0 | 4 |
| GYPA | SH2D3C | 0.668538 | 5 | 0 | 5 |
| GYPA | MMP19 | 0.667552 | 6 | 0 | 6 |
| GYPA | TMPRSS5 | 0.667093 | 6 | 0 | 6 |
| GYPA | KIR3DL3 | 0.665535 | 6 | 0 | 6 |
| GYPA | ARMC4 | 0.663416 | 5 | 0 | 5 |
| GYPA | PKMYT1 | 0.6626 | 3 | 0 | 3 |
For details and further investigation, click here