| Full name: KCNQ1 downstream neighbor | Alias Symbol: BWRT|HSA404617 | ||
| Type: non-coding RNA | Cytoband: 11p15.4 | ||
| Entrez ID: 55539 | HGNC ID: HGNC:13335 | Ensembl Gene: ENSG00000237941 | OMIM ID: 610980 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNQ1DN:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNQ1DN | 55539 | 220629_at | -0.1146 | 0.6008 | |
| GSE20347 | KCNQ1DN | 55539 | 220629_at | -0.0885 | 0.3133 | |
| GSE23400 | KCNQ1DN | 55539 | 220629_at | -0.2412 | 0.0000 | |
| GSE26886 | KCNQ1DN | 55539 | 220629_at | 0.1739 | 0.3068 | |
| GSE29001 | KCNQ1DN | 55539 | 220629_at | -0.1176 | 0.5358 | |
| GSE38129 | KCNQ1DN | 55539 | 220629_at | -0.2740 | 0.0223 | |
| GSE45670 | KCNQ1DN | 55539 | 220629_at | -0.0309 | 0.8131 | |
| GSE53622 | KCNQ1DN | 55539 | 13389 | 0.0086 | 0.9430 | |
| GSE53624 | KCNQ1DN | 55539 | 13389 | 0.2971 | 0.0014 | |
| GSE63941 | KCNQ1DN | 55539 | 220629_at | 0.1576 | 0.3959 | |
| GSE77861 | KCNQ1DN | 55539 | 220629_at | -0.0481 | 0.7005 |
Upregulated datasets: 0; Downregulated datasets: 0.
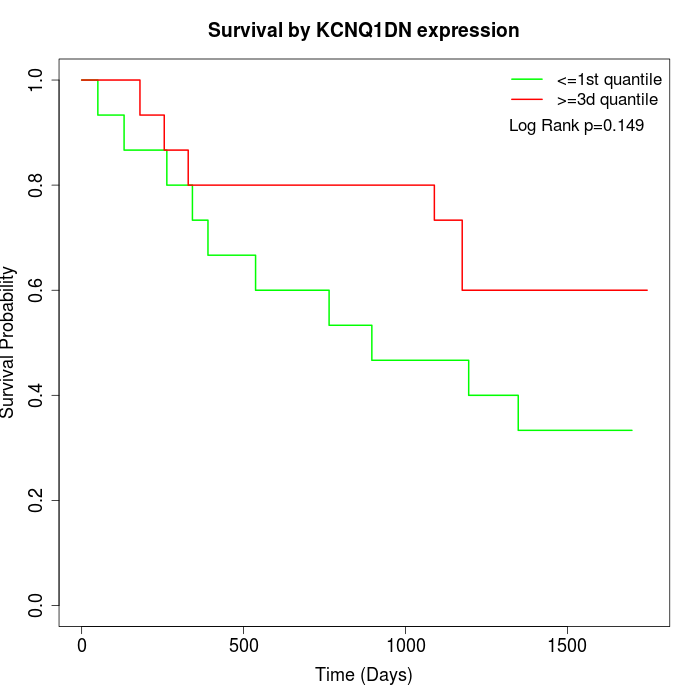
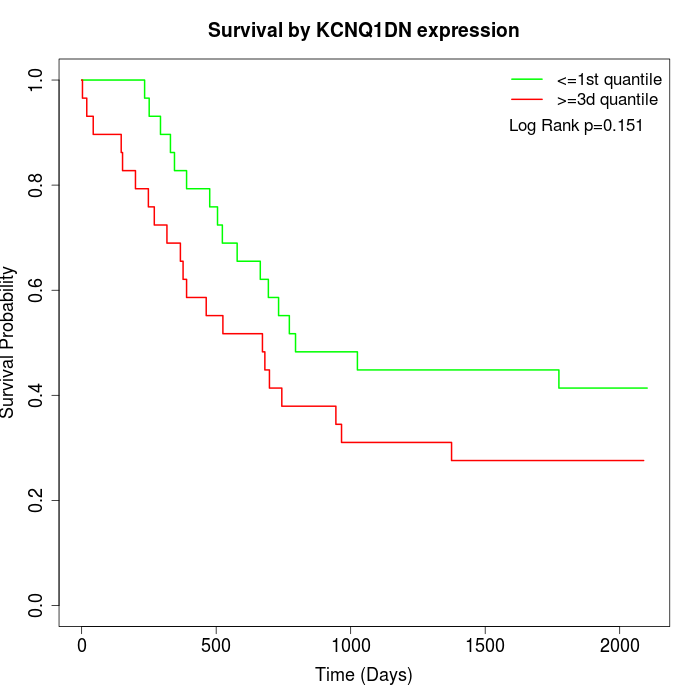
Survival by KCNQ1DN expression:
Note: Click image to view full size file.
Copy number change of KCNQ1DN:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNQ1DN | 55539 | 1 | 11 | 18 | |
| GSE20123 | KCNQ1DN | 55539 | 1 | 12 | 17 | |
| GSE43470 | KCNQ1DN | 55539 | 1 | 10 | 32 | |
| GSE46452 | KCNQ1DN | 55539 | 7 | 8 | 44 | |
| GSE47630 | KCNQ1DN | 55539 | 4 | 12 | 24 | |
| GSE54993 | KCNQ1DN | 55539 | 3 | 1 | 66 | |
| GSE54994 | KCNQ1DN | 55539 | 1 | 12 | 40 | |
| GSE60625 | KCNQ1DN | 55539 | 0 | 0 | 11 | |
| GSE74703 | KCNQ1DN | 55539 | 1 | 8 | 27 | |
| GSE74704 | KCNQ1DN | 55539 | 0 | 8 | 12 | |
| TCGA | KCNQ1DN | 55539 | 8 | 39 | 49 |
Total number of gains: 27; Total number of losses: 121; Total Number of normals: 340.
Somatic mutations of KCNQ1DN:
Generating mutation plots.
Highly correlated genes for KCNQ1DN:
Showing top 20/1210 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNQ1DN | IGH | 0.740796 | 6 | 0 | 6 |
| KCNQ1DN | GDF2 | 0.728371 | 8 | 0 | 8 |
| KCNQ1DN | MYEF2 | 0.718755 | 3 | 0 | 3 |
| KCNQ1DN | PCSK6 | 0.714768 | 4 | 0 | 4 |
| KCNQ1DN | SGCA | 0.713099 | 3 | 0 | 3 |
| KCNQ1DN | APBA1 | 0.70573 | 7 | 0 | 7 |
| KCNQ1DN | LINC00347 | 0.705443 | 3 | 0 | 3 |
| KCNQ1DN | RGR | 0.700551 | 6 | 0 | 6 |
| KCNQ1DN | PAPOLB | 0.698189 | 3 | 0 | 3 |
| KCNQ1DN | C11orf16 | 0.695076 | 8 | 0 | 8 |
| KCNQ1DN | LIMS2 | 0.692466 | 3 | 0 | 3 |
| KCNQ1DN | LIPF | 0.688806 | 3 | 0 | 3 |
| KCNQ1DN | AMN | 0.685721 | 8 | 0 | 8 |
| KCNQ1DN | CGA | 0.684711 | 5 | 0 | 5 |
| KCNQ1DN | MMP16 | 0.684133 | 4 | 0 | 4 |
| KCNQ1DN | TAAR5 | 0.678363 | 4 | 0 | 3 |
| KCNQ1DN | NPTXR | 0.677999 | 7 | 0 | 6 |
| KCNQ1DN | EPX | 0.676454 | 6 | 0 | 5 |
| KCNQ1DN | NYX | 0.675561 | 7 | 0 | 7 |
| KCNQ1DN | SSTR4 | 0.675057 | 7 | 0 | 6 |
For details and further investigation, click here