| Full name: acetyl-CoA acetyltransferase 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 6q25.3 | ||
| Entrez ID: 39 | HGNC ID: HGNC:94 | Ensembl Gene: ENSG00000120437 | OMIM ID: 100678 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of ACAT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ACAT2 | 39 | 209608_s_at | -0.2352 | 0.8774 | |
| GSE20347 | ACAT2 | 39 | 209608_s_at | 0.0011 | 0.9968 | |
| GSE23400 | ACAT2 | 39 | 209608_s_at | 0.0960 | 0.2376 | |
| GSE26886 | ACAT2 | 39 | 209608_s_at | -0.9770 | 0.0092 | |
| GSE29001 | ACAT2 | 39 | 209608_s_at | -0.3512 | 0.2894 | |
| GSE38129 | ACAT2 | 39 | 209608_s_at | 0.2279 | 0.2451 | |
| GSE45670 | ACAT2 | 39 | 209608_s_at | 0.0601 | 0.8206 | |
| GSE53622 | ACAT2 | 39 | 74710 | 0.2858 | 0.0025 | |
| GSE53624 | ACAT2 | 39 | 74710 | 0.5840 | 0.0000 | |
| GSE63941 | ACAT2 | 39 | 209608_s_at | 0.0097 | 0.9921 | |
| GSE77861 | ACAT2 | 39 | 209608_s_at | -0.2122 | 0.3323 | |
| GSE97050 | ACAT2 | 39 | A_23_P31135 | 0.1240 | 0.5502 | |
| SRP007169 | ACAT2 | 39 | RNAseq | -0.1668 | 0.7281 | |
| SRP008496 | ACAT2 | 39 | RNAseq | -0.4059 | 0.2266 | |
| SRP064894 | ACAT2 | 39 | RNAseq | -0.2346 | 0.4507 | |
| SRP133303 | ACAT2 | 39 | RNAseq | -0.4979 | 0.1764 | |
| SRP159526 | ACAT2 | 39 | RNAseq | -0.0464 | 0.8400 | |
| SRP193095 | ACAT2 | 39 | RNAseq | -0.4586 | 0.0047 | |
| SRP219564 | ACAT2 | 39 | RNAseq | -0.4948 | 0.5033 | |
| TCGA | ACAT2 | 39 | RNAseq | 0.2491 | 0.0374 |
Upregulated datasets: 0; Downregulated datasets: 0.
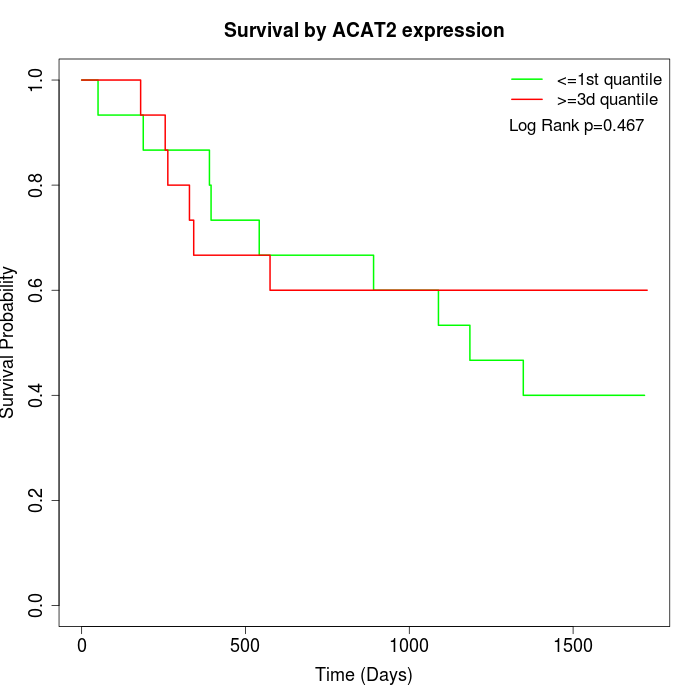
Survival by ACAT2 expression:
Note: Click image to view full size file.
Copy number change of ACAT2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ACAT2 | 39 | 1 | 5 | 24 | |
| GSE20123 | ACAT2 | 39 | 1 | 4 | 25 | |
| GSE43470 | ACAT2 | 39 | 4 | 0 | 39 | |
| GSE46452 | ACAT2 | 39 | 3 | 10 | 46 | |
| GSE47630 | ACAT2 | 39 | 9 | 4 | 27 | |
| GSE54993 | ACAT2 | 39 | 3 | 2 | 65 | |
| GSE54994 | ACAT2 | 39 | 8 | 8 | 37 | |
| GSE60625 | ACAT2 | 39 | 0 | 1 | 10 | |
| GSE74703 | ACAT2 | 39 | 4 | 0 | 32 | |
| GSE74704 | ACAT2 | 39 | 0 | 2 | 18 | |
| TCGA | ACAT2 | 39 | 13 | 22 | 61 |
Total number of gains: 46; Total number of losses: 58; Total Number of normals: 384.
Somatic mutations of ACAT2:
Generating mutation plots.
Highly correlated genes for ACAT2:
Showing top 20/96 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ACAT2 | JMJD8 | 0.820794 | 3 | 0 | 3 |
| ACAT2 | SHOC2 | 0.776917 | 3 | 0 | 3 |
| ACAT2 | TBL3 | 0.719487 | 3 | 0 | 3 |
| ACAT2 | HOGA1 | 0.717547 | 3 | 0 | 3 |
| ACAT2 | SLC35B4 | 0.703723 | 3 | 0 | 3 |
| ACAT2 | DENND2C | 0.682218 | 3 | 0 | 3 |
| ACAT2 | WDR44 | 0.676034 | 3 | 0 | 3 |
| ACAT2 | ATG9A | 0.669583 | 3 | 0 | 3 |
| ACAT2 | RHOV | 0.656866 | 3 | 0 | 3 |
| ACAT2 | COX8A | 0.656485 | 4 | 0 | 3 |
| ACAT2 | TRIM7 | 0.649039 | 4 | 0 | 4 |
| ACAT2 | FAM160A1 | 0.640811 | 3 | 0 | 3 |
| ACAT2 | TMEM45B | 0.634938 | 3 | 0 | 3 |
| ACAT2 | CORO2A | 0.632911 | 4 | 0 | 3 |
| ACAT2 | KATNBL1 | 0.630726 | 3 | 0 | 3 |
| ACAT2 | BTBD11 | 0.625199 | 3 | 0 | 3 |
| ACAT2 | NUP98 | 0.615078 | 4 | 0 | 3 |
| ACAT2 | EPHA1 | 0.613135 | 5 | 0 | 4 |
| ACAT2 | LY6G6C | 0.611089 | 4 | 0 | 3 |
| ACAT2 | PRSS2 | 0.607779 | 4 | 0 | 3 |
For details and further investigation, click here