| Full name: ADAM metallopeptidase domain 7 | Alias Symbol: GP-83|EAPI | ||
| Type: protein-coding gene | Cytoband: 8p21.2 | ||
| Entrez ID: 8756 | HGNC ID: HGNC:214 | Ensembl Gene: ENSG00000069206 | OMIM ID: 607310 |
| Drug and gene relationship at DGIdb | |||
Expression of ADAM7:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ADAM7 | 8756 | 211238_at | -0.0577 | 0.7873 | |
| GSE20347 | ADAM7 | 8756 | 211238_at | 0.1023 | 0.1250 | |
| GSE23400 | ADAM7 | 8756 | 211238_at | -0.0761 | 0.0064 | |
| GSE26886 | ADAM7 | 8756 | 211238_at | 0.0003 | 0.9978 | |
| GSE29001 | ADAM7 | 8756 | 211238_at | -0.0295 | 0.8207 | |
| GSE38129 | ADAM7 | 8756 | 211238_at | 0.0200 | 0.7642 | |
| GSE45670 | ADAM7 | 8756 | 211238_at | 0.0911 | 0.1766 | |
| GSE53622 | ADAM7 | 8756 | 198 | -0.2469 | 0.0027 | |
| GSE53624 | ADAM7 | 8756 | 198 | 0.1603 | 0.0861 | |
| GSE63941 | ADAM7 | 8756 | 239079_at | -0.0757 | 0.6551 | |
| GSE77861 | ADAM7 | 8756 | 239079_at | -0.0358 | 0.7609 |
Upregulated datasets: 0; Downregulated datasets: 0.
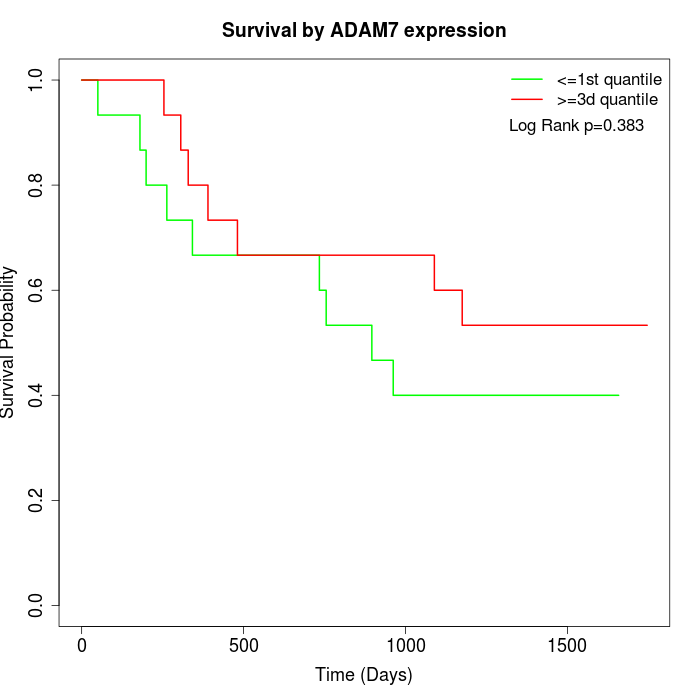
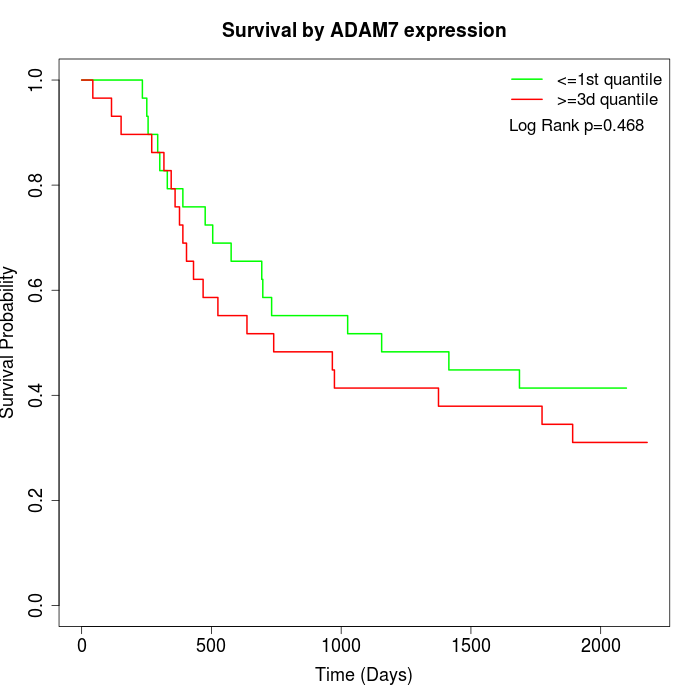
Survival by ADAM7 expression:
Note: Click image to view full size file.
Copy number change of ADAM7:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ADAM7 | 8756 | 4 | 12 | 14 | |
| GSE20123 | ADAM7 | 8756 | 4 | 12 | 14 | |
| GSE43470 | ADAM7 | 8756 | 4 | 7 | 32 | |
| GSE46452 | ADAM7 | 8756 | 13 | 13 | 33 | |
| GSE47630 | ADAM7 | 8756 | 10 | 8 | 22 | |
| GSE54993 | ADAM7 | 8756 | 2 | 14 | 54 | |
| GSE54994 | ADAM7 | 8756 | 8 | 18 | 27 | |
| GSE60625 | ADAM7 | 8756 | 3 | 0 | 8 | |
| GSE74703 | ADAM7 | 8756 | 4 | 6 | 26 | |
| GSE74704 | ADAM7 | 8756 | 3 | 8 | 9 | |
| TCGA | ADAM7 | 8756 | 15 | 43 | 38 |
Total number of gains: 70; Total number of losses: 141; Total Number of normals: 277.
Somatic mutations of ADAM7:
Generating mutation plots.
Highly correlated genes for ADAM7:
Showing top 20/456 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ADAM7 | OR1F1 | 0.708773 | 5 | 0 | 5 |
| ADAM7 | NAA11 | 0.702673 | 3 | 0 | 3 |
| ADAM7 | KCNA10 | 0.698106 | 3 | 0 | 3 |
| ADAM7 | SLC30A3 | 0.68413 | 4 | 0 | 4 |
| ADAM7 | SCGB1D1 | 0.678572 | 3 | 0 | 3 |
| ADAM7 | ALDOB | 0.675734 | 4 | 0 | 4 |
| ADAM7 | SLC9A3 | 0.675049 | 3 | 0 | 3 |
| ADAM7 | BCAN | 0.659637 | 4 | 0 | 4 |
| ADAM7 | CSN2 | 0.657548 | 3 | 0 | 3 |
| ADAM7 | CLDN16 | 0.654759 | 5 | 0 | 5 |
| ADAM7 | PDZD3 | 0.654544 | 4 | 0 | 4 |
| ADAM7 | KCNV1 | 0.653193 | 5 | 0 | 5 |
| ADAM7 | NTNG1 | 0.650235 | 4 | 0 | 4 |
| ADAM7 | CAMK2B | 0.647878 | 5 | 0 | 5 |
| ADAM7 | UMOD | 0.647167 | 5 | 0 | 4 |
| ADAM7 | IFNA7 | 0.646648 | 5 | 0 | 3 |
| ADAM7 | FCAR | 0.644716 | 4 | 0 | 4 |
| ADAM7 | TMPRSS5 | 0.643954 | 4 | 0 | 4 |
| ADAM7 | PRAMEF12 | 0.637992 | 5 | 0 | 5 |
| ADAM7 | GML | 0.637313 | 5 | 0 | 5 |
For details and further investigation, click here