| Full name: potassium voltage-gated channel modifier subfamily V member 1 | Alias Symbol: Kv8.1 | ||
| Type: protein-coding gene | Cytoband: 8q23.2 | ||
| Entrez ID: 27012 | HGNC ID: HGNC:18861 | Ensembl Gene: ENSG00000164794 | OMIM ID: 608164 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNV1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNV1 | 27012 | 215736_at | 0.0888 | 0.7192 | |
| GSE20347 | KCNV1 | 27012 | 215736_at | 0.1052 | 0.3564 | |
| GSE23400 | KCNV1 | 27012 | 215736_at | 0.0263 | 0.4762 | |
| GSE26886 | KCNV1 | 27012 | 215736_at | -0.1316 | 0.4176 | |
| GSE29001 | KCNV1 | 27012 | 215736_at | 0.0600 | 0.7635 | |
| GSE38129 | KCNV1 | 27012 | 215736_at | -0.0300 | 0.8050 | |
| GSE45670 | KCNV1 | 27012 | 215736_at | 0.1048 | 0.1837 | |
| GSE53622 | KCNV1 | 27012 | 54629 | -0.0120 | 0.9611 | |
| GSE53624 | KCNV1 | 27012 | 54629 | 0.5241 | 0.0003 | |
| GSE63941 | KCNV1 | 27012 | 215736_at | 0.0575 | 0.7094 | |
| GSE77861 | KCNV1 | 27012 | 215736_at | -0.0241 | 0.8118 | |
| SRP133303 | KCNV1 | 27012 | RNAseq | 0.1095 | 0.3276 | |
| SRP159526 | KCNV1 | 27012 | RNAseq | 0.8786 | 0.0363 | |
| SRP193095 | KCNV1 | 27012 | RNAseq | 0.1378 | 0.3493 | |
| SRP219564 | KCNV1 | 27012 | RNAseq | 0.1455 | 0.7146 | |
| TCGA | KCNV1 | 27012 | RNAseq | 1.1845 | 0.3318 |
Upregulated datasets: 0; Downregulated datasets: 0.
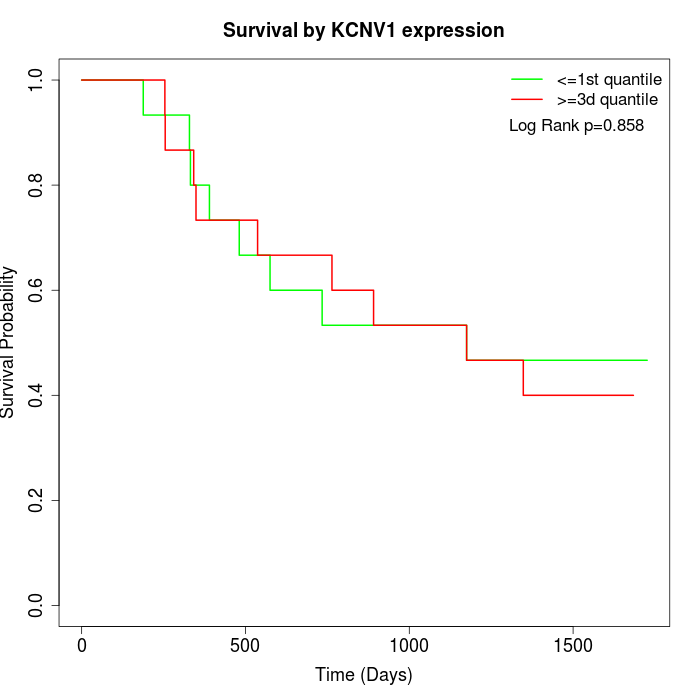
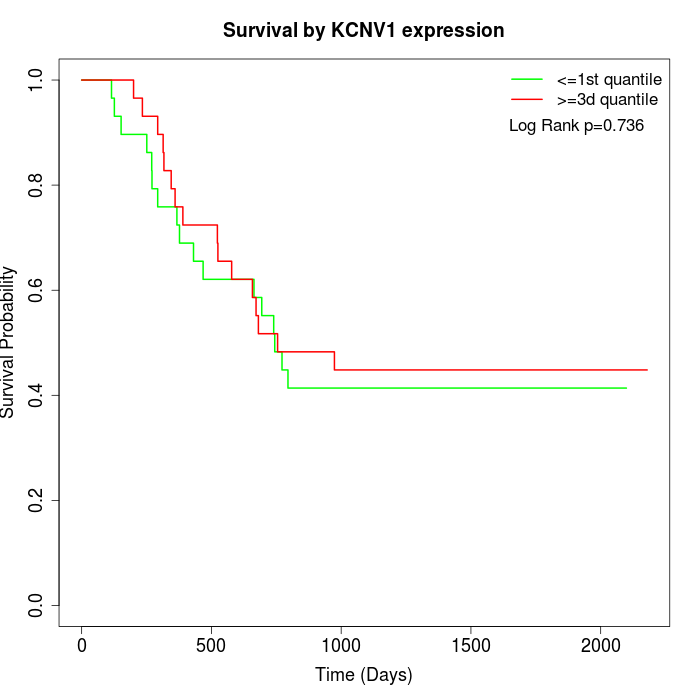
Survival by KCNV1 expression:
Note: Click image to view full size file.
Copy number change of KCNV1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNV1 | 27012 | 16 | 0 | 14 | |
| GSE20123 | KCNV1 | 27012 | 17 | 0 | 13 | |
| GSE43470 | KCNV1 | 27012 | 22 | 0 | 21 | |
| GSE46452 | KCNV1 | 27012 | 25 | 0 | 34 | |
| GSE47630 | KCNV1 | 27012 | 24 | 0 | 16 | |
| GSE54993 | KCNV1 | 27012 | 0 | 20 | 50 | |
| GSE54994 | KCNV1 | 27012 | 37 | 1 | 15 | |
| GSE60625 | KCNV1 | 27012 | 0 | 4 | 7 | |
| GSE74703 | KCNV1 | 27012 | 19 | 0 | 17 | |
| GSE74704 | KCNV1 | 27012 | 11 | 0 | 9 | |
| TCGA | KCNV1 | 27012 | 62 | 0 | 34 |
Total number of gains: 233; Total number of losses: 25; Total Number of normals: 230.
Somatic mutations of KCNV1:
Generating mutation plots.
Highly correlated genes for KCNV1:
Showing top 20/350 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNV1 | MED24 | 0.778696 | 3 | 0 | 3 |
| KCNV1 | ADAM22 | 0.750124 | 3 | 0 | 3 |
| KCNV1 | CA7 | 0.74988 | 3 | 0 | 3 |
| KCNV1 | CHST4 | 0.732812 | 3 | 0 | 3 |
| KCNV1 | CDSN | 0.72947 | 3 | 0 | 3 |
| KCNV1 | NMBR | 0.729318 | 3 | 0 | 3 |
| KCNV1 | PTPRJ | 0.719778 | 3 | 0 | 3 |
| KCNV1 | KCNJ14 | 0.718251 | 3 | 0 | 3 |
| KCNV1 | DAB1 | 0.7075 | 3 | 0 | 3 |
| KCNV1 | NTNG1 | 0.706277 | 3 | 0 | 3 |
| KCNV1 | INE1 | 0.705866 | 3 | 0 | 3 |
| KCNV1 | KIR3DL3 | 0.705734 | 5 | 0 | 5 |
| KCNV1 | PRKAR2A | 0.699557 | 3 | 0 | 3 |
| KCNV1 | RUNDC3A | 0.698315 | 4 | 0 | 3 |
| KCNV1 | CYP3A4 | 0.693407 | 3 | 0 | 3 |
| KCNV1 | ELAVL2 | 0.692644 | 4 | 0 | 4 |
| KCNV1 | IL21 | 0.688918 | 3 | 0 | 3 |
| KCNV1 | IL17A | 0.684998 | 3 | 0 | 3 |
| KCNV1 | HPX | 0.684804 | 5 | 0 | 4 |
| KCNV1 | PRPS1L1 | 0.684767 | 3 | 0 | 3 |
For details and further investigation, click here