| Full name: ATP/GTP binding protein like 4 | Alias Symbol: FLJ14442|CCP6 | ||
| Type: protein-coding gene | Cytoband: 1p33 | ||
| Entrez ID: 84871 | HGNC ID: HGNC:25892 | Ensembl Gene: ENSG00000186094 | OMIM ID: 616476 |
| Drug and gene relationship at DGIdb | |||
Expression of AGBL4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | AGBL4 | 84871 | 1553412_at | 0.0092 | 0.9790 | |
| GSE26886 | AGBL4 | 84871 | 1553412_at | 0.0201 | 0.9139 | |
| GSE45670 | AGBL4 | 84871 | 1553412_at | -0.0236 | 0.8304 | |
| GSE53622 | AGBL4 | 84871 | 6347 | -0.4236 | 0.0056 | |
| GSE53624 | AGBL4 | 84871 | 6347 | -0.1929 | 0.1151 | |
| GSE63941 | AGBL4 | 84871 | 1553412_at | -0.0131 | 0.9509 | |
| GSE77861 | AGBL4 | 84871 | 1553412_at | -0.1841 | 0.2204 | |
| GSE97050 | AGBL4 | 84871 | A_33_P3386572 | 0.0881 | 0.6756 | |
| SRP133303 | AGBL4 | 84871 | RNAseq | 0.5317 | 0.0094 | |
| TCGA | AGBL4 | 84871 | RNAseq | 0.4452 | 0.5618 |
Upregulated datasets: 0; Downregulated datasets: 0.
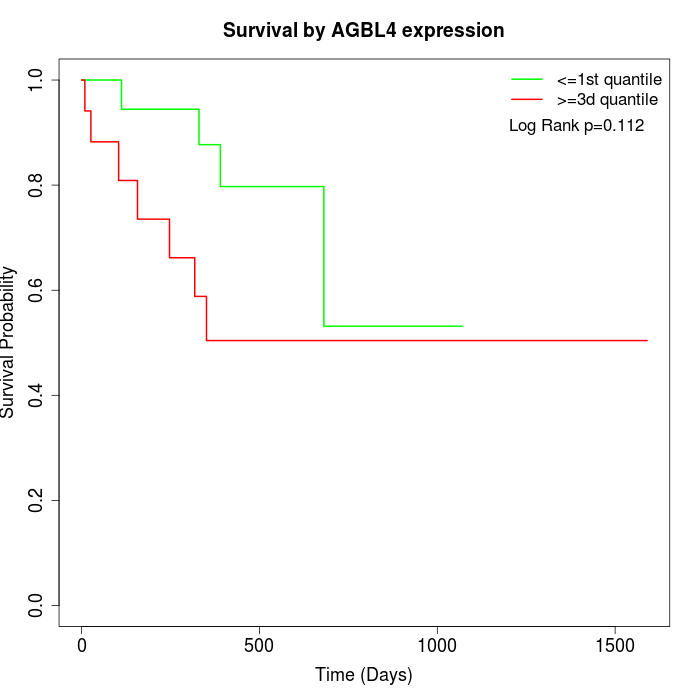
Survival by AGBL4 expression:
Note: Click image to view full size file.
Copy number change of AGBL4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | AGBL4 | 84871 | 3 | 3 | 24 | |
| GSE20123 | AGBL4 | 84871 | 3 | 2 | 25 | |
| GSE43470 | AGBL4 | 84871 | 5 | 4 | 34 | |
| GSE46452 | AGBL4 | 84871 | 2 | 1 | 56 | |
| GSE47630 | AGBL4 | 84871 | 8 | 4 | 28 | |
| GSE54993 | AGBL4 | 84871 | 0 | 1 | 69 | |
| GSE54994 | AGBL4 | 84871 | 11 | 2 | 40 | |
| GSE60625 | AGBL4 | 84871 | 0 | 0 | 11 | |
| GSE74703 | AGBL4 | 84871 | 4 | 2 | 30 | |
| GSE74704 | AGBL4 | 84871 | 2 | 0 | 18 | |
| TCGA | AGBL4 | 84871 | 10 | 18 | 68 |
Total number of gains: 48; Total number of losses: 37; Total Number of normals: 403.
Somatic mutations of AGBL4:
Generating mutation plots.
Highly correlated genes for AGBL4:
Showing top 20/47 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| AGBL4 | RPH3AL | 0.666893 | 3 | 0 | 3 |
| AGBL4 | SIGLEC1 | 0.664547 | 4 | 0 | 3 |
| AGBL4 | CMA1 | 0.661821 | 3 | 0 | 3 |
| AGBL4 | CELA2B | 0.632943 | 3 | 0 | 3 |
| AGBL4 | SPAG11A | 0.612555 | 4 | 0 | 3 |
| AGBL4 | SYNGAP1 | 0.609316 | 3 | 0 | 3 |
| AGBL4 | LINGO3 | 0.607818 | 4 | 0 | 4 |
| AGBL4 | LINC00470 | 0.5977 | 3 | 0 | 3 |
| AGBL4 | TMEM114 | 0.597692 | 4 | 0 | 4 |
| AGBL4 | SLC6A18 | 0.594971 | 3 | 0 | 3 |
| AGBL4 | RBP5 | 0.574959 | 4 | 0 | 3 |
| AGBL4 | CSH1 | 0.572505 | 5 | 0 | 3 |
| AGBL4 | TSGA10IP | 0.572164 | 5 | 0 | 5 |
| AGBL4 | ARMC5 | 0.571573 | 3 | 0 | 3 |
| AGBL4 | WFDC8 | 0.570445 | 5 | 0 | 5 |
| AGBL4 | TAS2R3 | 0.570079 | 4 | 0 | 3 |
| AGBL4 | CECR9 | 0.566598 | 3 | 0 | 3 |
| AGBL4 | TTLL3 | 0.566055 | 4 | 0 | 3 |
| AGBL4 | BSND | 0.559981 | 6 | 0 | 4 |
| AGBL4 | NR2E3 | 0.556203 | 3 | 0 | 3 |
For details and further investigation, click here