| Full name: Rho GTPase activating protein 5 | Alias Symbol: RhoGAP5|p190-B|p190BRhoGAP | ||
| Type: protein-coding gene | Cytoband: 14q12 | ||
| Entrez ID: 394 | HGNC ID: HGNC:675 | Ensembl Gene: ENSG00000100852 | OMIM ID: 602680 |
| Drug and gene relationship at DGIdb | |||
ARHGAP5 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04510 | Focal adhesion | |
| hsa04670 | Leukocyte transendothelial migration |
Expression of ARHGAP5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ARHGAP5 | 394 | 233849_s_at | -0.2532 | 0.7951 | |
| GSE20347 | ARHGAP5 | 394 | 217936_at | -0.7328 | 0.0033 | |
| GSE23400 | ARHGAP5 | 394 | 217936_at | -0.5027 | 0.0000 | |
| GSE26886 | ARHGAP5 | 394 | 233849_s_at | -0.9330 | 0.0009 | |
| GSE29001 | ARHGAP5 | 394 | 217936_at | -0.9639 | 0.0093 | |
| GSE38129 | ARHGAP5 | 394 | 217936_at | -0.6217 | 0.0018 | |
| GSE45670 | ARHGAP5 | 394 | 217936_at | -0.4632 | 0.0020 | |
| GSE53622 | ARHGAP5 | 394 | 121558 | -0.4417 | 0.0000 | |
| GSE53624 | ARHGAP5 | 394 | 121558 | -0.6422 | 0.0000 | |
| GSE63941 | ARHGAP5 | 394 | 233849_s_at | 0.2979 | 0.6785 | |
| GSE77861 | ARHGAP5 | 394 | 233849_s_at | -0.4939 | 0.3246 | |
| GSE97050 | ARHGAP5 | 394 | A_33_P3386459 | -0.3208 | 0.2707 | |
| SRP007169 | ARHGAP5 | 394 | RNAseq | -1.1077 | 0.0005 | |
| SRP008496 | ARHGAP5 | 394 | RNAseq | -0.9452 | 0.0000 | |
| SRP064894 | ARHGAP5 | 394 | RNAseq | -1.1257 | 0.0010 | |
| SRP133303 | ARHGAP5 | 394 | RNAseq | -0.3060 | 0.1974 | |
| SRP159526 | ARHGAP5 | 394 | RNAseq | -0.8695 | 0.0098 | |
| SRP193095 | ARHGAP5 | 394 | RNAseq | -0.8049 | 0.0000 | |
| SRP219564 | ARHGAP5 | 394 | RNAseq | -0.7808 | 0.1293 | |
| TCGA | ARHGAP5 | 394 | RNAseq | -0.0601 | 0.3539 |
Upregulated datasets: 0; Downregulated datasets: 2.
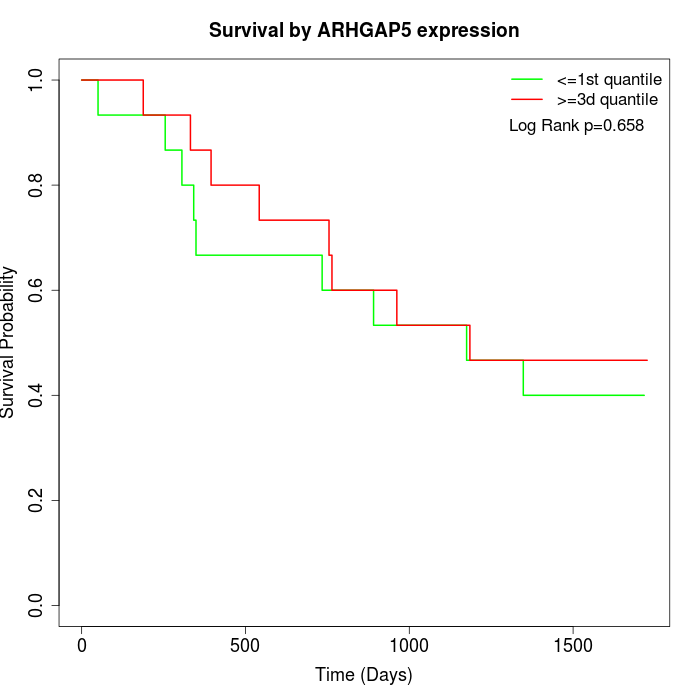
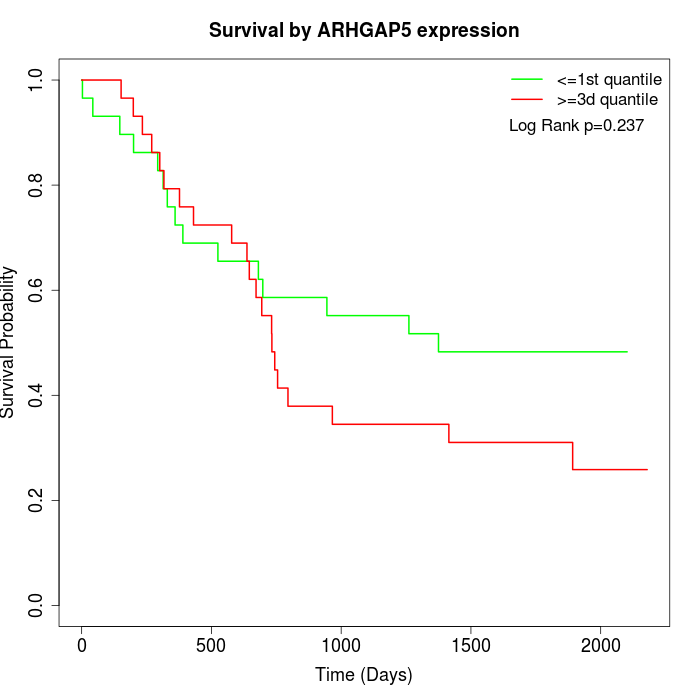
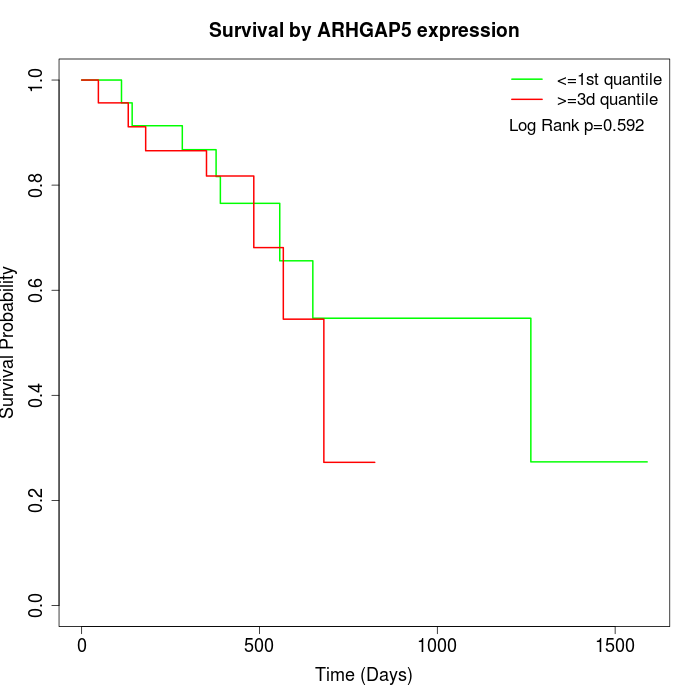
Survival by ARHGAP5 expression:
Note: Click image to view full size file.
Copy number change of ARHGAP5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ARHGAP5 | 394 | 10 | 4 | 16 | |
| GSE20123 | ARHGAP5 | 394 | 10 | 4 | 16 | |
| GSE43470 | ARHGAP5 | 394 | 10 | 2 | 31 | |
| GSE46452 | ARHGAP5 | 394 | 17 | 2 | 40 | |
| GSE47630 | ARHGAP5 | 394 | 11 | 10 | 19 | |
| GSE54993 | ARHGAP5 | 394 | 3 | 11 | 56 | |
| GSE54994 | ARHGAP5 | 394 | 19 | 5 | 29 | |
| GSE60625 | ARHGAP5 | 394 | 0 | 2 | 9 | |
| GSE74703 | ARHGAP5 | 394 | 9 | 2 | 25 | |
| GSE74704 | ARHGAP5 | 394 | 4 | 3 | 13 | |
| TCGA | ARHGAP5 | 394 | 31 | 16 | 49 |
Total number of gains: 124; Total number of losses: 61; Total Number of normals: 303.
Somatic mutations of ARHGAP5:
Generating mutation plots.
Highly correlated genes for ARHGAP5:
Showing top 20/745 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ARHGAP5 | MAP4K3 | 0.749697 | 3 | 0 | 3 |
| ARHGAP5 | NAA35 | 0.726047 | 4 | 0 | 4 |
| ARHGAP5 | LYSMD3 | 0.718399 | 3 | 0 | 3 |
| ARHGAP5 | GTF2B | 0.7141 | 3 | 0 | 3 |
| ARHGAP5 | TRIM23 | 0.713347 | 3 | 0 | 3 |
| ARHGAP5 | ZNF155 | 0.708951 | 3 | 0 | 3 |
| ARHGAP5 | MIPOL1 | 0.698738 | 3 | 0 | 3 |
| ARHGAP5 | METTL14 | 0.695122 | 3 | 0 | 3 |
| ARHGAP5 | GGA3 | 0.683931 | 3 | 0 | 3 |
| ARHGAP5 | AP5M1 | 0.681662 | 5 | 0 | 5 |
| ARHGAP5 | ARHGAP5-AS1 | 0.678583 | 7 | 0 | 6 |
| ARHGAP5 | ZCCHC9 | 0.67847 | 3 | 0 | 3 |
| ARHGAP5 | HEATR5A | 0.668419 | 7 | 0 | 6 |
| ARHGAP5 | CLK1 | 0.668354 | 4 | 0 | 3 |
| ARHGAP5 | MTMR3 | 0.664751 | 9 | 0 | 8 |
| ARHGAP5 | ZNF514 | 0.664434 | 3 | 0 | 3 |
| ARHGAP5 | MRPL46 | 0.663661 | 3 | 0 | 3 |
| ARHGAP5 | DCTN4 | 0.658032 | 4 | 0 | 3 |
| ARHGAP5 | RWDD2B | 0.655898 | 4 | 0 | 3 |
| ARHGAP5 | ZNF585B | 0.655702 | 3 | 0 | 3 |
For details and further investigation, click here