| Full name: aryl hydrocarbon receptor nuclear translocator like | Alias Symbol: MOP3|JAP3|BMAL1|PASD3|bHLHe5 | ||
| Type: protein-coding gene | Cytoband: 11p15.3 | ||
| Entrez ID: 406 | HGNC ID: HGNC:701 | Ensembl Gene: ENSG00000133794 | OMIM ID: 602550 |
| Drug and gene relationship at DGIdb | |||
ARNTL involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04710 | Circadian rhythm | |
| hsa04728 | Dopaminergic synapse |
Expression of ARNTL:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ARNTL | 406 | 209824_s_at | 0.0196 | 0.9790 | |
| GSE20347 | ARNTL | 406 | 209824_s_at | 0.0549 | 0.7998 | |
| GSE23400 | ARNTL | 406 | 209824_s_at | 0.0208 | 0.7338 | |
| GSE26886 | ARNTL | 406 | 209824_s_at | 1.0579 | 0.0018 | |
| GSE29001 | ARNTL | 406 | 209824_s_at | 0.1443 | 0.6718 | |
| GSE38129 | ARNTL | 406 | 209824_s_at | -0.0257 | 0.9028 | |
| GSE45670 | ARNTL | 406 | 209824_s_at | 0.3116 | 0.4029 | |
| GSE53622 | ARNTL | 406 | 30108 | 0.1570 | 0.2901 | |
| GSE53624 | ARNTL | 406 | 90060 | 0.2303 | 0.0208 | |
| GSE63941 | ARNTL | 406 | 209824_s_at | -0.6425 | 0.3384 | |
| GSE77861 | ARNTL | 406 | 209824_s_at | 0.3196 | 0.2505 | |
| GSE97050 | ARNTL | 406 | A_33_P3385567 | -0.1006 | 0.7117 | |
| SRP007169 | ARNTL | 406 | RNAseq | -0.0414 | 0.9405 | |
| SRP008496 | ARNTL | 406 | RNAseq | 0.2435 | 0.4748 | |
| SRP064894 | ARNTL | 406 | RNAseq | 0.1053 | 0.7022 | |
| SRP133303 | ARNTL | 406 | RNAseq | 0.1469 | 0.5551 | |
| SRP159526 | ARNTL | 406 | RNAseq | -0.7656 | 0.0534 | |
| SRP193095 | ARNTL | 406 | RNAseq | 0.3150 | 0.0538 | |
| SRP219564 | ARNTL | 406 | RNAseq | -0.2426 | 0.7349 | |
| TCGA | ARNTL | 406 | RNAseq | 0.0575 | 0.4554 |
Upregulated datasets: 1; Downregulated datasets: 0.
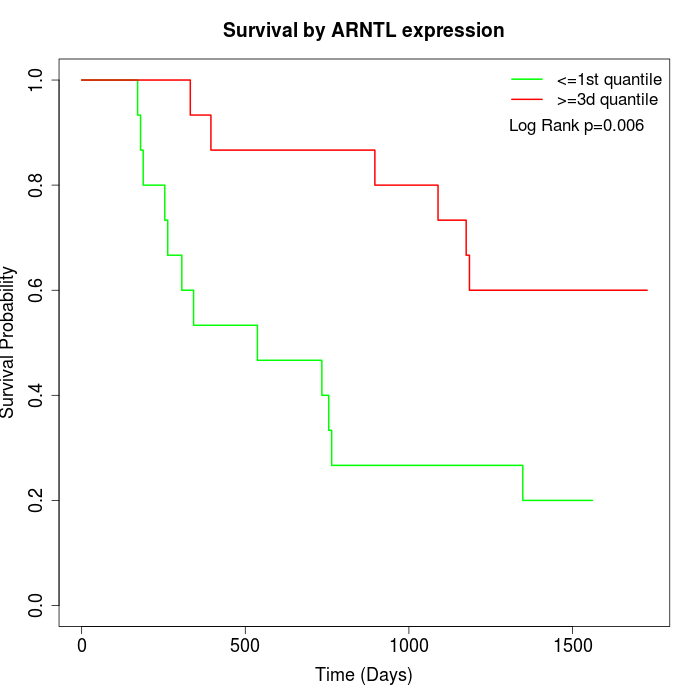
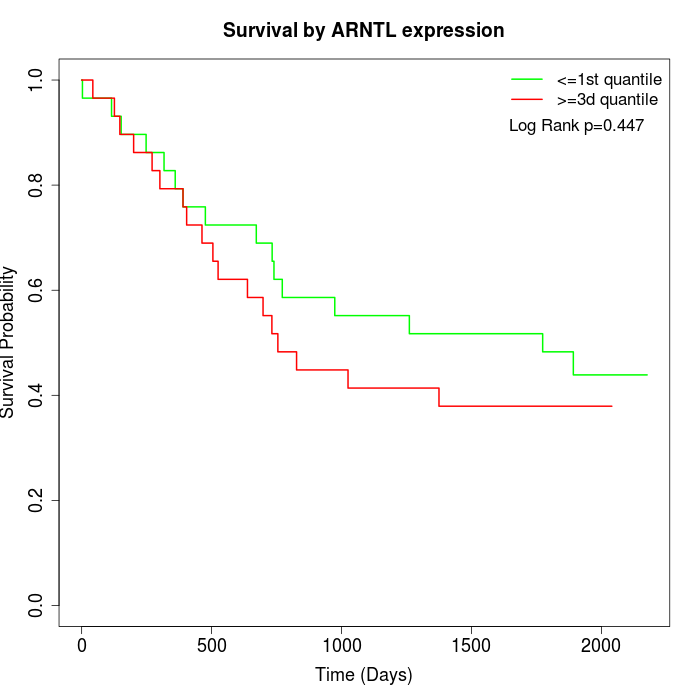
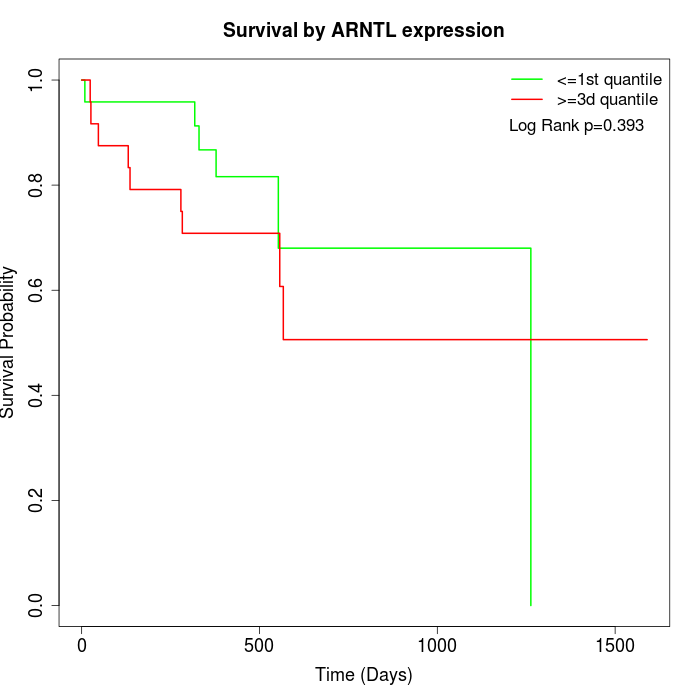
Survival by ARNTL expression:
Note: Click image to view full size file.
Copy number change of ARNTL:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ARNTL | 406 | 0 | 9 | 21 | |
| GSE20123 | ARNTL | 406 | 0 | 9 | 21 | |
| GSE43470 | ARNTL | 406 | 1 | 5 | 37 | |
| GSE46452 | ARNTL | 406 | 7 | 5 | 47 | |
| GSE47630 | ARNTL | 406 | 3 | 11 | 26 | |
| GSE54993 | ARNTL | 406 | 3 | 1 | 66 | |
| GSE54994 | ARNTL | 406 | 4 | 12 | 37 | |
| GSE60625 | ARNTL | 406 | 0 | 0 | 11 | |
| GSE74703 | ARNTL | 406 | 1 | 3 | 32 | |
| GSE74704 | ARNTL | 406 | 0 | 8 | 12 | |
| TCGA | ARNTL | 406 | 13 | 31 | 52 |
Total number of gains: 32; Total number of losses: 94; Total Number of normals: 362.
Somatic mutations of ARNTL:
Generating mutation plots.
Highly correlated genes for ARNTL:
Showing all 16 correlated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ARNTL | HEATR4 | 0.687608 | 3 | 0 | 3 |
| ARNTL | CCPG1 | 0.680038 | 3 | 0 | 3 |
| ARNTL | BPTF | 0.602608 | 3 | 0 | 3 |
| ARNTL | MAP2 | 0.600983 | 3 | 0 | 3 |
| ARNTL | RBBP8 | 0.586332 | 3 | 0 | 3 |
| ARNTL | ODC1 | 0.58616 | 5 | 0 | 3 |
| ARNTL | NKD2 | 0.576226 | 4 | 0 | 3 |
| ARNTL | BLOC1S6 | 0.562963 | 6 | 0 | 4 |
| ARNTL | RMDN1 | 0.56223 | 3 | 0 | 3 |
| ARNTL | HOMER1 | 0.547803 | 6 | 0 | 3 |
| ARNTL | RPS15A | 0.538866 | 4 | 0 | 3 |
| ARNTL | LONP1 | 0.52885 | 4 | 0 | 3 |
| ARNTL | PRMT3 | 0.523428 | 9 | 0 | 6 |
| ARNTL | MLF1 | 0.522118 | 5 | 0 | 3 |
| ARNTL | DENND4A | 0.509504 | 5 | 0 | 3 |
| ARNTL | ETAA1 | 0.500236 | 6 | 0 | 3 |
For details and further investigation, click here